Automatically creating pivot table column names in PostgreSQL
Often in bioinformatics I receive a dataset that is entirely non-relational. For instance, every row is a gene, every column is a biological sample, and the cell values are the expression levels of each gene measured by microarray. To join such datasets to others (e.g. metadata on the samples and on the genes), I need to relationalize the table. In the above example, that would mean building a new table with three columns (gene, sample, expression_level). Trouble is, then later if I want to do any fancy matrix math on the data I need to de-relationalize it, getting back to the rows=genes, columns=samples layout. This is called a pivot table and can be achieved in PostgreSQL using the crosstab() function, but there’s a catch: you need to type out all of the column names.
To avoid spending my life typing out column names, I wrote a function in Postgres procedural language (PL/pgSQL) that will generate a crosstab query automatically. You can’t execute the generated crosstab query automatically because PL/pgSQL functions that return tables (setof record functions) cannot have the number and type of columns determined on the fly. Instead this function returns the query in a varchar, and then you can execute the query to get the pivot table you want. That’s an extra round-trip to the database if you’re working programmatically, or an extra copy-and-paste if you’re working in the query editor. But it’s better than typing out column names.
Here’s the code. I run PostgreSQL 9.2 but I believe this should work at least as far back as 8.4.
-- PL/pgSQL code to create pivot tables with automatic column names
-- Eric Minikel, CureFFI.org - 2013-03-19
-- prerequisite: install the tablefunc module
create extension tablefunc;
-- tablename: name of source table you want to pivot
-- rowc: the name of the column in source table you want to be the rows
-- colc: the name of the column in source table you want to be the columns
-- cellc: an aggregate expression determining how the cell values will be created
-- celldatatype: desired data type for the cells
create or replace function pivotcode (tablename varchar, rowc varchar, colc varchar, cellc varchar, celldatatype varchar) returns varchar language plpgsql as $$
declare
dynsql1 varchar;
dynsql2 varchar;
columnlist varchar;
begin
-- 1. retrieve list of column names.
dynsql1 = 'select string_agg(distinct ''_''||'||colc||'||'' '||celldatatype||''','','' order by ''_''||'||colc||'||'' '||celldatatype||''') from '||tablename||';';
execute dynsql1 into columnlist;
-- 2. set up the crosstab query
dynsql2 = 'select * from crosstab (
''select '||rowc||','||colc||','||cellc||' from '||tablename||' group by 1,2 order by 1,2'',
''select distinct '||colc||' from '||tablename||' order by 1''
)
as newtable (
'||rowc||' varchar,'||columnlist||'
);';
return dynsql2;
end
$$
-- toy example to show how it works
create table table_to_pivot (
rowname varchar,
colname varchar,
cellval numeric
);
insert into table_to_pivot values ('row1','col1',11);
insert into table_to_pivot values ('row1','col2',12);
insert into table_to_pivot values ('row1','col3',13);
insert into table_to_pivot values ('row2','col1',21);
insert into table_to_pivot values ('row2','col2',22);
insert into table_to_pivot values ('row2','col3',23);
insert into table_to_pivot values ('row3','col1',31);
insert into table_to_pivot values ('row3','col2',32);
insert into table_to_pivot values ('row3','col3',33);
select pivotcode('table_to_pivot','rowname','colname','max(cellval)','integer');
Executing that last select statement will return the following:
select * from crosstab (
'select rowname,colname,max(cellval) from table_to_pivot group by 1,2 order by 1,2',
'select distinct colname from table_to_pivot order by 1'
)
as newtable (
rowname varchar,_col1 integer,_col2 integer,_col3 integer
);
And executing that query will in turn give you:
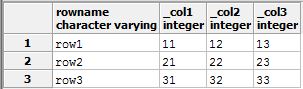
The underscore in front of the column names is so that it will still work if the columns start with numbers.
When I tried this on some of my real datasets, it worked great when the columns were, say, chromosomes – anything that numbers in the tens. When I tried to do it on a set with hundreds of patient samples as the columns, my columnlist parameter came back empty. I’m not sure why that is: varchar is the same as text, and text‘s maximum size is 1 GB, and no particular limit is specified for string_agg itself. But even if I could fix that, one could easily bump up against the maximum number of columns in Postgres, which is apparently 250-1600 depending on column types.
There are some other solutions to this problem on the web as well – see this one using crosstab_hash, or since I’m often doing the downstream analysis in R anyway, I could use melt and cast on the R side. But there have been a few times I thought it would be useful to crosstab directly in SQL. Hopefully you’ll find this useful as well.
