PrP transgene constructs
Background on vectors for transgenesis
Now that I’ve learned a bit more about cloning and various types of vectors in my introductory genetics class I sat down to acquaint myself with the different types of clones of the prion protein gene that have been used in transgenic animals.
First off, here is a quick review of the sizes of different types of vectors that people use, as reviewed in Lodish 2000 Chapter 7.2, particularly Table 7.2, and Strachan & Reed 1999 Chapter 4 (Table 4.2):
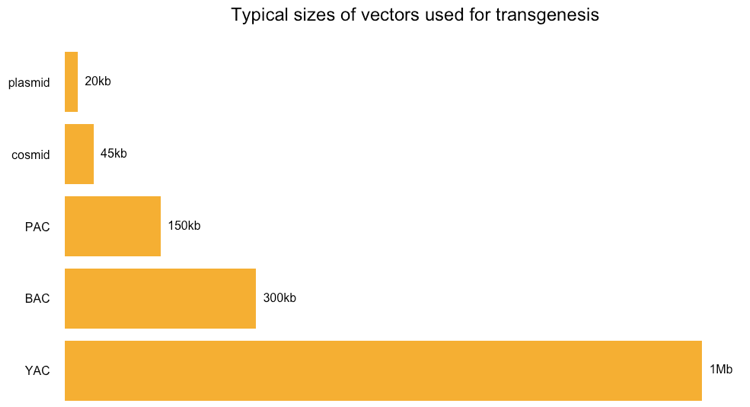
Most of these things are just different kinds of plasmids. Above and in the cited references, “plasmid” by itself refers to standard high copy number plasmids, which have relatively low capacity. Cosmids are special plasmids containing a phage cos sequence and able to accept a larger insert of foreign DNA. P1 phage artificial chromosomes (PACs) are packaged into P1 bacteriphages but circularize once they are adsorbed into a bacterial cell, so they too may be thought of as plasmids. Bacterial artificial chromosomes (BACs) are just very large low copy number plasmids, usually based on the F factor. Of the above options, only yeast artificial chromosomes (YACs) are fundamentally different, as they require producing your DNA in yeast instead of bacteria.
To my understanding, the larger the vector, the more difficult it is to work with. If you’re doing transgenesis, then you’re microinjecting DNA into pronuclei as explained here, and the larger vectors are more likely to shear apart upon being forced through the tiny needle. That’s one motivation for picking the smallest vector that gets the job done. But what “the job” is here is debatable - do you want the smallest construct that makes a mouse sick, or do you want to include the entire genomic locus in an effort to as closely mimic physiological expression patterns as possible? Efforts to model Huntington’s disease, for instance, have spawned a wide spectrum of mouse models, from R6/2 with their tiny single-exon transgene [Mangiarini 1996], N171-82Q with their two-exon transgene [Schilling 1999], Hdh mice with mutant human exon 1 knocked in [Wheeler 1999], to BACHD [Gray 2008] and YAC128 mice [Hodgson 1999] with the entire human HTT locus.
Unsuccessful attempts at PrP expression via cDNA clones
As far as I can tell, the first PrP transgenes were created from Syrian hamster sequence and were reported by Prusiner’s group in [Scott 1989]. In hamsters, as in humans, Prnp has two exons, with the entire coding sequence in the second one. The first construct reported in this paper is a minigene dubbed migHaPrP containing a 1.5kb piece of the hamster PrP promoter, a 1.7kb hamster cDNA and a 0.8 fragment containing the polyadenylation signal, for a total of ~4kb. They were only able to obtain one line of mice harboring the migHaPrP transgene, but these mice were not susceptible to hamster prions (Table 1) and their PrP expression was not characterized in Figs 1-2.
Similarly, Weissmann’s group created a transgene with a mouse cDNA plus 6kb of 5’ sequence containing the prion promoter and 2.2kb of 3’ sequence presumably containing the polyadenylation signal [Fischer 1996]. In mice, Prnp has three exons, the first two of which are just 5’UTR and the last of which contains the whole coding sequence - so of course a cDNA lacks both introns, fusing all three exons together. This ~10kb construct didn’t work:
in all eight lines tested, even in those… with very high transgene copy numbers (>150 copies), showed no detectable levels of PrP protein in the brain and only very weak (tgb24/+) or no expression of PrP RNA
— [Fischer 1996]
So, cDNAs appear not to be sufficient to drive PrP expression.
Successful PrP transgenes
cos-Tet
That first paper from Prusiner’s group [Scott 1989] also reported the creation of a Syrian hamster cosmid clone containing the entire ~15kb hamster Prnp genomic sequence plus 24kb to the 5’ and 6kb to the 3’, so about 45kb total. This transgene achieved efficient expression in several transgenic mouse lines (Tg69, 71, 81). These and another line created with the same cosmid (Tg7) formed the basis of subsequent transgenesis experiments from the Prusiner lab [Prusiner 1990]. This construct, variously called the cos.Tet or cos-Tet vector, continues to be the basis of most of the newer transgenic lines from the Prusiner lab. The Tg4053 mouse, which overexpresses mouse PrP-A, has the Prnpa open reading frame in the cos-Tet vector [Carlson 1994]. Many other species’ open reading frames, or chimeras thereof, have likewise been swapped into the protein-coding portion of this cosmid [e.g. Giles 2010, Watts 2012]. Rocky Mountain Labs and MRC Prion Unit have both adopted cos-Tet for their mouse models as well [e.g. Race 2009, Asante 2009].
I/LnJ cosmid
Westaway and Prusiner also created a separate cosmid clone of the mouse Prnpb allele from I/LnJ mice [Westaway 1991]. That cosmid contains the entire Prnp gene plus 6kb of 5’ sequence and 15.5kb of 3’ sequence, and Westaway’s group eventually found that it also includes Prnd, which encodes Doppel [Moore 1999]. The mouse reference genome, which is based on Prnpa mice, has the Prnp transcript as spanning 28.5kb (chr2:131909928-131938436 in mm10), but the Prnpa allele turns out to contain a 6.5kb retroviral insertion of an intracisternal A-particle (IAP) sequence in intron 2 which is absent in Prnpb [Lee 1998]. Therefore I’d expect the Prnpb cosmid to be 6kb + (28.5-6.5)kb + 15.5kb = 43.5kb; it is reported to be in fact only 39.76kb in total [Moore 1999]. I’m not sure what accounts for this small discrepancy.
Half-genomic construct
Weissmann and Aguzzi had similar luck creating transgenic mice using larger cosmid clones. They created one line of mice using Westaway’s Prnpb construct, and several lines using a new “half-genomic” (HG) construct they created [Fischer 1996]. This construct was created by fusing together 5.5kb of 5’ promoter sequence and 1.3kb of mouse exon 1, intron 1 and exon 2 from Westaway’s Prnpb clone to 1.3kb of a MoPrP-A cDNA and 3kb of 3’ flanking sequence from a Prnpa mouse. All told, this construct weighs in at about 12kb. The sacrifice of mouse intron 2 makes it considerably smaller than the 45kb cosmid clones, though it lacks PrP expression in Purkinje cells. Case Western has adopted the half-genomic construct for its transgenic lines [Kong 2005]. In addition, two different lines of mice have been created which express just a smaller, 5kb fragment of the “half-genomic” construct under a doxycycline-controlled “Tet-off” (tTA) [Tremblay 1998] or “Tet-on” (rtTA) [Huang 2007] promoter. These lines both exhibit toxic ectopic and/or heterochronic expression of PrP.
Human PRNP in a P1 artificial chromosome
Vincent Beringue and Hubert Laude at France’s INRA created a transgenic line (tg650) expressing HuPrP 129M which utilizes the entire human PRNP locus [Beringue 2008]. They used a P1 artificial chromosome containing 110kb of human sequence including PRNP as well as PRND and possibly other neighboring genes, and microinjected it into Zurich I PrP knockout mice [Bueler 1992]. They obtained one founder with 5 copies of this PAC in cis, and bred it to homozygosity for a total of 10 copies, achieving ~6x wild-type expression levels. The transmission properties of CJD and absence of spontaneous disease in these mice have been well characterized [Beringue 2008]. Because these mice include the entire human sequence and not just the human open reading frame, they would be especially valuable for gene therapy studies where you might want to direct siRNA, ASOs or CRISPRs against non-coding sequence in PRNP.
Summary
Any attempt to compare the above transgenes side-by-side is by necessity fanciful, since the mapping between hamster, mouse and human is not perfect. Nonetheless, I wanted some sort of visual aid to see at a glance which regions of sequence were included in which construct. This is the best representation I’ve come up with:
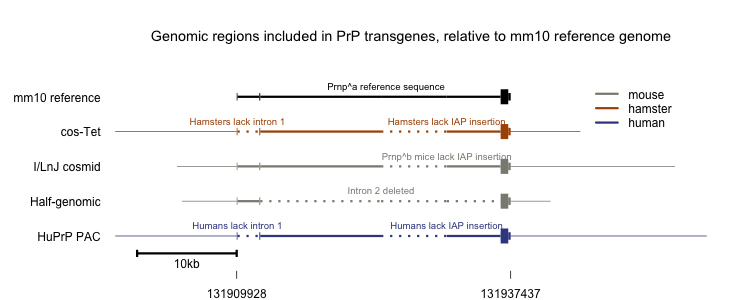
I didn’t find any other transgenes in use in the literature - if you know of any others, let me know.
