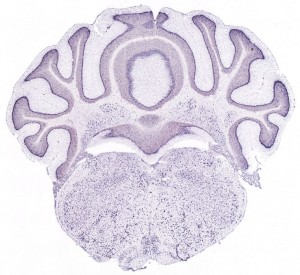
Mouse brain images from the Allen Institute for Brain Science
Thinking more about my post yesterday on machine learning for image segmentation, I’m excited to implement a machine learning approach to figuring out how to recognize astrocytes. At a minimum, I could automate the trying out of all the different options in CellProfiler, which would already be helpful and might well find something that works. And I could also try implementing a search over convolutional networks (CNs) like Jain et al describe. And hopefully in the process I’ll learn a lot about machine learning, about image segmentation and gain some more familiarity with CellProfiler in particular.
One challenge is I don’t yet have access to a large volume of high-resolution images of prion-diseased and healthy brain tissue. I need an image set to practice on and get started learning this stuff. Yesterday I was forwarded a link to some data hosted at the Allen Institute for Brain Science, which self-describes as “A growing collection of online public resources integrating extensive gene expression and neuroanatomical data, complete with a novel suite of search and viewing tools.” For instance I think what I’m looking at here is a rotatable 3D map of where PRNP is expressed in the mouse brain. It looks like they may also have the kind of image data I’m looking for. These I think are cross sectional images of where PRNP is expressed (I’m still not clear how these images were taken– is this with GFP swapped in at the PRNP locus??) On the web the images are all hidden in composites where you can only download the current field of view — to download whole images at high resolution you have to go through their API. After a bit of fiddling I was able to find the images for PRNP expression through this series of requests:
Search for PRNP
http://mouse.brain-map.org/aba/api/gene/search?term=PRNP
Get imageseries list for PRNP
http://mouse.brain-map.org/aba/api/gene/Prnp.xml
Get list of images in each imageseries for PRNP
http://mouse.brain-map.org/aba/api/imageseries/1405.xml
http://mouse.brain-map.org/aba/api/imageseries/1406.xml
Get any particular image (whole image, full resolution) from the imageseries (just swap in &path= any downloadImagePath from the xml docs above):
http://mouse.brain-map.org/aba/api/image?zoom=-1&path=/external/aibssan/production4/Prnp_Baylor_7917/zoomify/primary/null/Prnp_44_null_B.aff
Easy enough–that actually got me a whole 4MB image. Here’s what these images look like (this is a lower res version):
It’s a jpg, which is a little odd– aren’t images from microscopy usually stored in a lossless format? In any event, the quality is quite good. I still need to figure out what I’m looking at though. This seems to be a whole brain, in which case I would think the little dots are too big to be individual cells?