Improvements in GATK 2.x
Yesterday I attended a lecture by the folks at the Broad Institute about the new release of Genome Analysis Toolkit (GATK) 2.x. For those who don’t know, GATK (together with BWA and Picard, with which it comes bundled) is an end-to-end solution for sequencing, going from raw FASTQ reads coming off of a sequencer all the way to finished genotypes or haplotypes.
I have been working on a pipeline for analyzing exome data and, after many software problems, had initially given up on GATK and used an alternative approach (blog post forthcoming). However this workshop has given me enough reason to go back and try again to get GATK working on my data. First, it’s emerging as a gold standard: apparently it now boasts 6,000 users, making it far and away the most widely used solution. Second, improvements are coming along fast, and that is much of what this talk was about. Here are a few highlights of 2.x as opposed to 1.x:
- Reduced reads. GATK can take a BAM file and retain the full diversity of original reads only in places where the reads disagree with each other; for any bases where all (actually the default value is 95% or more) of the reads agree with each other (whether or not they agree with the reference sequence), it compresses them down to one read with metadata indicating how many reads there originally were. This results in 20x – 100x compression, though obviously only on deep coverage files (any regions with 1x coverage obviously can’t be compressed at all). This means it’s easier to move files around, and that variant calling will be several times faster.
- Base quality recalibration has been improved. Base quality scores from sequencing companies tend to be inflated– ex. a PHRED score of 20 means 99% accuracy, yet in practice it is found that more than 1% of bases with score 20 are actually wrong. GATK can recalibrate the scores to make them more accurate (though still not perfect).
- Combined SNP, indel and structural variant calling. This is all in one step instead of separate steps.
- Better indel calling. Indels are still a lot harder to call than SNPs and more of them fail validation, but it’s improving.
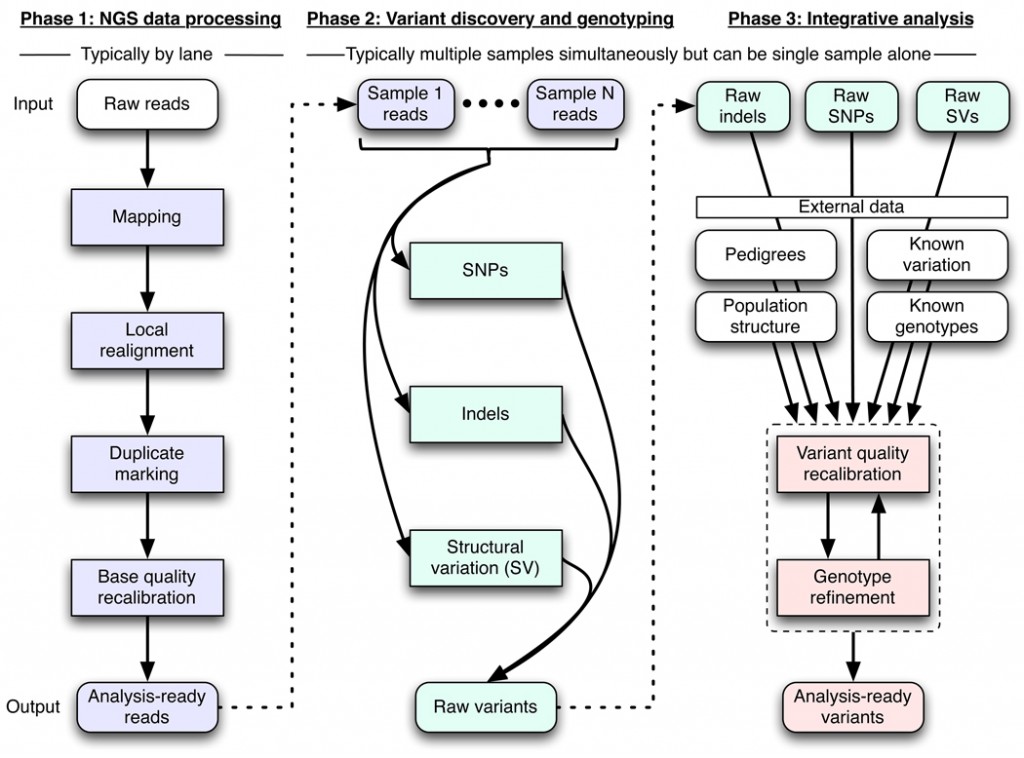
Another highlight was something I believe is not actually new, but that I didn’t know about before: Queue. There are a LOT of steps in GATK. To wit:
That’s Figure 1 from DePristo 2011‘s paper on GATK. So naturally it becomes a pain having to pipeline all these steps yourself, and deal with job scheduling, memory requirements, intermediate files, etc. Queue is supposed to take care of that for you. They also gave us an order of magnitude estimate of time savings which is always helpful — a UnifiedGenotyper job which would take 10 days to run in series can be parallelized to run in 6 hours using Queue.