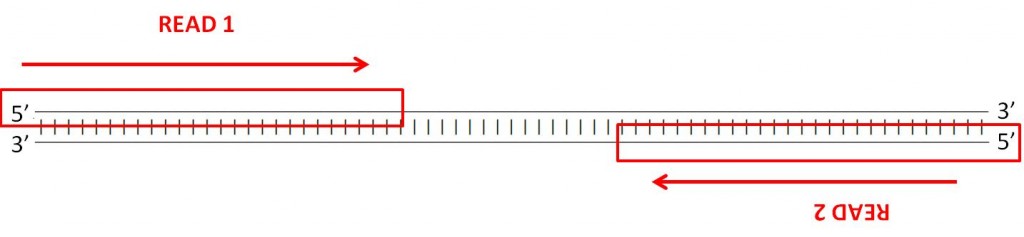
Forward and reverse reads in paired-end sequencing
This topic is incredibly easy to get confused about, so here is as clear an explanation as I can muster. It will start out big picture and then get into the weeds.
Sequencing by synthesis, which is how most commercially available high-throughput sequencing technologies work as of December 2012 (see notes on sequencing technologies), always synthesizes the new strand (which becomes your read) in a 5′-to-3′ direction. That’s because this is how DNA polymerase works in our cells (indeed, in every living thing’s cells) and sequencing relies on DNA polymerase. Since the new strand is synthesized 5′-to-3′, you are working your way up the template strand in a 3′-to-5′ direction.
In any sequencing technology, you PCR amplify the individual DNA fragments once they have hybridized to flowcells or beads. This means you end up with both strands of DNA. If you were to read both of the strands from their respective 3′ ends at once, you’d be getting two different sequences and your results would be uninterpretable. To avoid this problem, sequencing technologies ligate non-complementary adapters to the 3′ and 5′ ends of DNA fragments so that the primer for one adapter only begins synthesis on one strand and not on its complement.
In conventional paired-end sequencing, you simply sequence using the adapter for one end, and then once you’re done you start over sequencing using the adapter for the other end.
This means your two reads are the reverse complement of the 100 3′-most bases of the Watson strand and the Crick strand; these reads are assumed to be identical to the 100 5′-most bases of the Crick strand and Watson strand respectively.
Here’s what your reads represent, then:
Therefore when you open your FASTQ files and look at a pair of reads, the sequences you see are, conceptually, pointing towards each other on opposite strands. When you align them to the genome, one read should align to the forward strand, and the other should align to the reverse strand, at a higher base pair position than the first one so that they are pointed towards one another. This is known as an “FR” read – forward/reverse, in that order.
This is all for conventional paired-end sequencing. Some specialized technologies, such as using circularized DNA fragments to create large insert jumping libraries [Talkowski 2011], switch things around so that your reads ought to align in an “RF” position – reverse/forward, in that order. This is different from FR because it means the reverse read aligned at a lower base pair position than the forward read, and thus that they are pointing away from another.
But if you’re just doing conventional paired-end sequencing (i.e. Illumina), your reads are supposed to align FR, and if they instead align RF, FF or RR, that’s a problem and often indicates the reads aligned incorrectly (though it could also mean they aligned correctly and that a real inversion or translocation exists in the sample’s genome – see notes from Devin Absher’s talk on calling structural variants). If read pairs don’t align FR, most aligners will flag them as “not a proper pair” in the SAM/BAM file by zeroing the FLAG 0×02 bit (proper pair flag) (see SAM spec). Heng Li, author of BWA, states here that BWA will only set the ‘proper pair flag’ to 1 for Illumina reads aligned FR (for SOLiD it allows FF or RR).
Therefore if you look at a SAM/BAM file (for Illumina data at least), it should be the case that in any pair of reads with the 0×02 bit set (i.e. considered a proper pair), exactly one of the two reads will have the 0×10 bit set as well (i.e. it is reverse-complemented; again, see the SAM file spec). For the read with its 0×10 bit set, the “SEQ” listed in the SAM file will be the reverse complement of the original read as seen in the FASTQ. That means that in the SAM file, the SEQs for a pair of reads are now both being presented in forward orientation even though the “FR” orientation information is stored in the FLAG.
For reads that don’t form a proper pair, or aren’t mapped at all, (almost) all bets are off. Pairs might be FF, RR or RF, and one might be mapped and the other not. Moreover, an unmapped read might have the 0×10 flag set, or not.
Why bother reverse-complementing the read if it doesn’t align anywhere anyway? I don’t know; I can’t find any information in the BWA documentation about why this might occur. But I spot checked the FLAGs of unmapped reads in one of my BAMs:
cut -f 2 1_unmapped_sorted.sam | sort | uniq
And found that a variety of FLAG values occur: [101,103,117,133,141,151,157,165,167,181,69,77,87], several of which have the 0×10 bit set, for instance 117. When I go back and pull out a sampling of the reads with flag value 117:
grep $'\t117\t' 1_unmapped_sorted.sam | head | less
And then compare the reads in those to my original FASTQs, I find that they are indeed reverse complemented, for instance:
grep $'\t117\t' 1_unmapped_sorted.sam | head | less FCC0CHTACXX:6:1101:10003:55579#GCCAATAT 117 chr2 130057568 0 * = 130057568 0 GGGACACACTGAGCTCAGGGATAGGGTGGAGGTGGACTGGACTGAGAGCAGCGTCAGAGGGGAAGGCACTGCAGCAGGGGCCCGACATAGGCAGGGGTAC grep FCC0CHTACXX:6:1101:10003:55579 -m 1 -A 3 1_1.fq @FCC0CHTACXX:6:1101:10003:55579#GCCAATAT/1 GTACCCCTGCCTATGTCGGGCCCCTGCTGCAGTGCCTTCCCCTCTGACGCTGCTCTCAGTCCAGTCCACCTCCACCCTATCCCTGAGCTCAGTGTGTCCC + _b_eeeeegggggiihiiiiiiiiihhfhhiifhhifffhihhiiihhhfhghdgdg`cdeeeR]bdddbcccbbcca^bccc`bbccccccbcd_`b_b
(You’ll notice that RNAME for this read is chr2, but remember from the SAM spec that if 0×04 is set, no assumptions can be made about RNAME).
I conclude that even if 0×04 is set, it is still safe to assume that for any individual read that has its 0×10 flag set, the “SEQ” shown in the BAM file is actually the reverse complement of the original read from the FASTQ.
This came up because recently I was iterating through BAMs using pysam, trying to re-align unmapped reads, and for my particular purpose I wanted to have both of their sequences in the same orientation, i.e. both forward or both reverse. This was confusing at first because zero, one or both of them might already be reverse-complemented in the SEQ field. The most conceptually straightforward way is just to reverse complement whichever (neither, one or both) have .is_reverse = True, so that now you’re back to baseline, and then you reverse complement exactly one of them. To avoid the extra step, though, logically you could use an XNOR, so in Python:
if(read.is_reverse == mate.is_reverse): read = reversecomplement(read)
Addendum: for BWA, at least, the proper pair flag depends not only on the FR orientation but also on insert size being within a certain range (from BWA manual):
The maximum distance x for a pair considered to be properly paired (SAM flag 0×2) is calculated by solving equation Phi((x-mu)/sigma)=x/L*p0, where mu is the mean, sigma is the standard error of the insert size distribution, L is the length of the genome, p0 is prior of anomalous pair and Phi() is the standard cumulative distribution function. For mapping Illumina short-insert reads to the human genome, x is about 6-7 sigma away from the mean. Quartiles, mean, variance and x will be printed to the standard error output.