Cell Biology 04: The Secretory Pathway
These are notes from lecture 4 of Harvard Extension’s Cell Biology course.
The secretory pathway refers to the endoplasmic reticulum, Golgi apparatus and the vesicles that travel in between them as well as the cell membrane and lysosomes. It’s named ‘secretory’ for being the pathway by which the cell secretes proteins into the extracellular environment. But as usual, etymology only tells a fraction of the story. This pathway also processes proteins that will be membrane-bound (whether in the cellular membrane or in the ER or Golgi membranes themselves), as well as lysosomal enzymes, and also any proteins that will live their lives in the secretory pathway itself. It also does some things other than process proteins.
The cytosol and the ‘lumen’ (the liquid that fills the secretory pathway) are different chemical environments, and they normally never mix. The cytosol is reductive (when you’re in the cytosol, you keep meeting molecules that want to offer you electrons), and the ER, Golgi and extracellular environment are oxidative (molecules keep coming up to you asking for electrons). See redox if still confused. This makes for different protein-folding conditions: for instance, disulfide bonds usually only form in oxidative conditions. Moreover, different proteins may live only in the secretory pathway or only in the cytosol. The secretory pathway provides a route for the cell to handle things that might not be good to have in the cytoplasm, and/or are most useful when kept concentrated in a specialized compartment with their desired interacting partners. Hepatocytes (in the liver) sequester drugs and toxins in the smooth ER and break them down for excretion from the body there. The secretory pathway is not contiguous, but every movement between its components is in little bubbled-off microcosms of its own chemical world, called vesicles.
Many proteins that go through the secretory pathway never touch the cytosol – except the parts of membrane proteins that stick out on the cytosolic side. Many of them need chaperones to help with folding, and/or a whole series of post-translational modifications in order to be ready for their native function, and the secretory pathway specializes in providing them all of that.
Today’s lecture will focus on how proteins get translated into the ER and how they travel (in vesicles) between the ER, Golgi and other destinations. This is beautifully depicted in the Life of the Cell video:
The endoplasmic reticulum is the first step in the secretory pathway. Its membrane is continuous with the outer nuclear membrane, though it’s not clear why that matters, since it’s not like proteins begin their life in the nucleus. Rather, mRNAs drift around in the cytoplasm until they get picked up by a ribosome interested in translating them. In ‘posttranslational translocation’ the new protein is moved into the ER after it’s translated. In the more interesting phenomenon called ‘cotranslational translocation’ the ribosome starts translation just like any other protein, but somewhere in the first 16 to 30 amino acids it hits a signal peptide (aka signal sequence). That signal’s motif is often 1 positively charged amino acid followed by 6-12 hydrophobic amino acids. This motif gets recognized by signal recognition particle (SRP, a ‘ribonucleoprotein’ or hybrid RNA/protein molecule) which binds to it and prevents the ribosome from continuing translation. Translation is stopped until the ribosome/SRP complex encounters an SRP receptor on the ER membrane. When they meet, SRP and its receptor each bind one GTP molecule in the ER membrane, which apparently strengthens their interaction. Fortuitously, this all happens adjacent to a Sec61 translocon – a protein complex that forms a channel crossing the ER membrane. The translocon is actually a complex of three different proteins (genes: SEC61A1 or SEC61A2, SEC61B, SEC61G), of which the Sec61a subunit has 10 membrane-spanning a-helices which form the channel. Once the ribosome is docked at the membrane it continues translation, pushing the signal peptide and eventually the whole protein through the channel into the ER lumen. When translation stops, SRP and SRP receptor both hydrolzye their GTP to release each other and the ribosome cargo (this has to require the energy of GTP, since the original binding was downhill), a signal peptidase cleaves the signal peptide off of the nascent protein, and the protein is free to start folding in the ER.
A couple of other players are involved for some ER proteins. Oligosaccharide transferase, which adds glycosyl groups to asparagines in the nascent protein, is part of the translocon complex and it actually performs glycosylation while the new protein is still being translated. So although we call glycosylation a ‘post-translational modification’ it is actually made during translation in this case. Also, to achieve their proper structure, some proteins need to be fully translated before they are allowed to start folding – if the N-terminal portion was allowed to start folding as soon as it entered the lumen, it would end up with the wrong overall structure. To prevent this, sometimes BiP the chaperone binds the protein to keep it unfolded for a while. Imagine BiP as another Pac-Man that bites down on the protein to keep it linear, like Hsc70 in the mitochondrial targeting process (see last week).
Here’s a video of it:
The first couple of minutes show the basic scenario described above. Then it moves on to a more complex scenario I’ll introduce in a minute. FYI, the video depicts two ‘controversial’ things not included in the above description: (1) the signal peptide being degraded in the membrane, and (2) a ‘plug protein’ that stops up the channel before/after translation. Not all scientists agree on these two things yet.
All of the proteins that we know go through the secretory pathway were pinpointed there by people doing localization experiments to see where in the cell a protein lies. A weird fact about the ER is that you can put the cell in a blender and afterwards the ER will just start reconnecting to itself, forming little ‘microsomes’ that are not attached to the nucleus but form contiguous bubbles of ER. You can then start to play games with proteases – which break down proteins – and detergents – which solubilize the ER membrane. Assuming your protein of interest is translated, you can check if it (1) survives protease treatment but (2) doesn’t survive protease + detergent treatment, then it’s a secretory pathway protein. The logic is that in case (1) it was protected inside the ER, but in case (2) you dissolved the ER, so it got eaten by the protease. All this assumes you have an antibody or some other way of detecting whether the protein of interest is there after these treatments.
People also used such techniques to figure out that only 70 amino acids of a new protein can be translated before it becomes too late for that protein to end up in the ER. Remember, the signal peptide is in the first 16-30 amino acids, and translocation to the ER depends on SRP being present. Ribosomes translate at a predictable rate, so people got ribosomes started on translating some mRNA and then waited set amounts of time before adding SRP, to see how much translation could occur before SRP could no longer do its job.
The SRP receptor and the Sec61 proteins are ER membrane proteins – and there many other ER membrane, Golgi membrane and lysosome membrane proteins as well. In fact, even the membrane proteins (see class 02) of the cell membrane get processed in the secretory pathway. Many of these have several or tens of transmembrane domains (20-25 hydrophobic amino acids each) that have to be inserted in the correct order and orientation (for example, you really want your ion channels and transporters pointed in the right direction, into vs. out of the cell). Accordingly there are a bunch of fancy biological mechanisms for getting these proteins inserted into the membrane correctly. This is what the latter half of the above video depicts.
So here’s a tautology: some proteins have a topogenic sequence which determines their orientation in the membrane. This sequence is made of two types of signal sequences:
- a stop-transfer sequence (abbreviated STA for some reason) is a 22-25 hydrophobic amino acid sequence somewhere in the middle of the protein that forms an alpha helix. When encountered it gets shoved into the membrane, and then translation of the rest of the protein continues in the cytosol. So this kind of ‘undoes’ the translocation to the ER that was started by the signal peptide at the beginning (N terminus) of the protein.
- a signal anchor sequence (abbreviated SA) is also a 22-25aa hydrophobic alpha helix, but with a series of ~3 positively charged amino acids on its left or right. Like the signal peptide, this is recognized by SRP, which brings the ribosome to the ER. But unlike the signal peptide, this alpha helical sequence will be inserted into the ER membrane. The orientation of insertion is determined by the 3 positively charged amino acids. The positive charges have to always end up on the cytosolic side, so if they come after (i.e. C-terminal of) the hydrophobic sequence, the protein ends up with its C terminal end pointed into the cytosol, but if they come before (i.e. N-terminal of) the hydrophobic sequence, the protein ends up with its N terminus pointed into the cytosol.
With those two signals as building blocks, you can imagine a protein with a series of stop transfer and signal anchor sequences to create a whole series of back and forth transmembrane domains stitched into the membrane as if by a sewing machine. People have classified the membrane proteins into five categories:
- Type I has just a signal peptide and then one stop transfer in the middle. Therefore it ends up with its (hydrophilic) N terminus in the lumen, its (hydrophobic) middle in the membrane and its (hydrophilic) C terminus in the cytosol.
- Type II does not start with a signal peptide. It starts out like any other protein, but in the middle it has a signal anchor sequence with the +++ amino acids coming first and the hydrophobic series after. This makes the protein get translocated midway through translation, with the already-translated N-terminal part sticking out into the cytosol (since the +++ have to stay cytosolic) and the now-beginning-to-be-translated C-terminal part getting translated directly into the ER. So it ends up transmembrane with its C terminus in the ER and N terminus in the cytosol – opposite of Type I.
- Type III is like Type II – no signal peptide, just a signal anchor in the middle, but in this case the +++ come after the hydrophobic sequence, which reverses the orientation. So this ends up with its N terminus in the ER and its C terminus in the cytosol. Opposite of Type II and, in the end, the same as Type I, though it got there in a different way – it does not have a signal peptide that gets cleaved off in the ER.
- Type IV or ’multipass’ proteins have an alternating series of signal sequences and stop transfer sequences. These are clearly more than one ‘type’, yet are not nearly as diverse as your combinatoric imagination might allow. The orientation of the first signal sequence determines whether the N terminus will end up in the cytosol or ER, and total number of stop transfer + signal anchor sequences determines where the C terminus will end up: an even number = same side as N terminus, odd number = opposite side as N terminus. The STA and SA sequences have to strictly alternate, with the exception that you can start with two signal anchor sequences if the first one is oriented with the N terminus into the cytosol. Just to make a mockery of this categorization scheme, people have defined some incompletely-defined subtypes of Type IV, where Type IVa is N-terminal in cytosol (thus it starts like a Type II protein) and Type IVb is N-terminal in the lumen (it starts like a Type III protein but then has another SA sequence that puts it back into the ER). GLUT1 from Class 02 is a Type IVa.
- GPI-anchored proteins, which are the fifth type but aren’t called Type V, start with a signal peptide and end with a hydrophobic C-terminus which stays embedded in the membrane. That hydrophobic end gets cleaved off and replaced with GPI, which also stays embedded in the membrane. PrP is one of these – more on that later.
By now we’ve discussed how proteins can end up in the ER lumen or spanning the ER membrane. Most proteins leave the ER within minutes, transported in vesicles bound for the Golgi and then later for excretion, lysosomes or the cell membrane. That forward direction of travel is called anterograde; going backwards from Golgi to ER is retrograde transport.
Both types of transport take place in membrane-bound vesicles. These bud off of the membrane of wherever they’re coming from, and later fuse to the membrane of wherever they’re headed – beautifully depicted at ~2:25 in the Life of the Cell video above. The body from which the vesicles form is the ‘donor compartment’, and the destination they later fuse to is the ‘acceptor compartment’.
The budding process requires that G proteins in the membrane recruit Coat proteins. Specifically, for anterograde transport, G protein Sar1 (gene: SAR1A) recruits COPII (‘cop two’); for retrograde transport, an ARF G protein recruits COPI (pronounced ‘cop one’). These G proteins are activated to do this job when GEF loads them with GTP, swapping out GDP.
So the steps in anterograde transport, for example, are as follows:
- Sec12-GEF (Sec stands for secretory) loads Sar1 with GTP. When bound to GDP, Sar1 just floats around the donor compartment, but when bound to GTP, it undergoes conformational change that causes its otherwise-buried N-terminal hydrophobic tail to protrude, making it stick into the membrane, where COPII proteins then start to accumulate because they really like that tail.
- The COPIIs start to polymerize and, due to its conformation, have an intrinsic preference for curvature, so their accumulation starts to make budding happen. At the same time, membrane bound proteins that need to be transported – identified by a DXE (i.e. aspartate-anything-glutamate) amino acid sequence that forms a binding site in their cytosolic part – get recruited to the newly forming vesicle. Membrane-bound proteins act as receptors, recruiting lumenal proteins that are bound for the Golgi to hang out in the concave space where they’ll end up in the vesicle once it forms.
- Once enough COPII have arrived, the vesicle buds off, at which point Sar1 hydrolyzes its GTP, providing the energy for it to suck its hydrophobic tail back into itself, cutting the COPIIs loose. The vesicle is now disconnected from the donor compartment.
- Now, for poorly explained (or poorly understood?) reasons, the coat of COPIIs just disassembles, exposing receptors under the coat which direct the targeting of the vesicle. Once the vesicle arrives at its destination, Rab-GTP embedded in the vesicle membrane interacts with a Rab effector embedded in the acceptor compartment membrane. A sideways glance is exchanged, interest is kindled. Soon the vesicle will fuse to the membrane.
- SNARE proteins present on both the vesicle and target membrane (V-SNARE and T-SNARE respectively) interact to bring the membranes even closer. In this example we’ll consider VAMP (the VAMP_ genes) as the V-SNARE and Syntaxin (the STX__ genes) and SNAP25 (SNAP25 gene) as the T-SNAREs. Syntaxin and SNAP25 are both membrane proteins; Syntaxin has 1 alpha helix and SNAP25 has 2, all on the cytosolic side. The alpha helices drive the interaction with VAMP. The opposing sides’ alpha helices have extremely strong affinity for one another, bringing the membranes close enough to fuse. Once this has happened, prying the V-SNAREs and T-SNAREs apart again requires two proteins: NSF (gene: NSF; stands for NEM sensitive factor) and alpha-SNAP (gene: NAPA), a soluble NSF attachment protein. NSF is an ATPase, and burns ATP to drive the energetically uphill disassembly of the complex.
Now for retrograde transport. Why is there retrograde transport at all? Here is a non-exhaustive list of some reasons:
- Some membrane proteins start their life in the ER, need to get modified in the Golgi, but then need to get back to the ER. They do this with a KKXX amino acid sequence.
- There’s also a KDEL amino acid sequence at the C terminus of some lumenal proteins which is suppsoed to keep them in the ER, but it’s not perfect – sometimes they end up in the Golgi, in which case they’re targeted back to the ER via retrograde transport dependent on that KDEL sequence for recognition. The mechanism is kind of neat – the proteins that recognize and bind to KDEL do so only at low pH, and the pH of the Golgi is lower than the ER, so they bind KDEL in the Golgi, then release it when they’re back in the more neutral pH of the ER.
- Also, think about it, all the proteins that participate in anterograde transport – the V-SNARES, Rab, etc. – have to get back to the ER so they can do it all over again, like how the bus has to get back to the bus depot at the end of the day.
- As we’ll see shortly, the Golgi come in multiple stages which depend on the addition of enzymes from further downstream.
The process of retrograde transport is not so different from anterograde. It uses ARF instead of Sar1, COPI instead of COPII, but it works the same: ARF loaded with GTP lets its hydrophobic tail stick into the membrane, attracting the attention of COPIs. COPI has two components, COPIalpha and COPIbeta, both of which interact with that KKXXX sequence to recruit membrane-bound proteins destined for retrograde transport. Some proteins also have an RR sequence (anywhere in the protein) which can flag them for retrograde transport.
The Golgi apparatus is not contiguous. It is a stacked set of separate subcompartments called sacs or cisternae. Different compartments have different properties and proteins visit them in a particular order. In order from ER to cell membrane, the Golgi compartments are called cis, medial, trans and trans-Golgi network. Each compartment has different enzymes that modify proteins, and the modifications have to happen in a certain order, hence the need for a stacked set of compartments.
But as proteins mature in the Golgi, it’s not as though they bud off in vesicles from one compartment and move to the next. Rather, the compartment they are already in moves outward and ‘matures’ as new enzymes are added to it (from further down the Golgi chain) via retrograde transport. Weird, right? It’s kind of like if instead of moving from an elementary school to a middle school to a high school you just stayed in one school building for your whole childhood and adolescence, and they just brought in new textbooks and teachers every year to keep it appropriate to the grade that you and your classmates had now reached. Here’s what the Golgi look like as they move and evolve:
So there’s (little or) no anterograde transport within the Golgi, but plenty of retrograde transport to bring each new round of enzymes in. When proteins have finally completed the full K-12 curriculum of the Golgi network, they do undergo transport to move on to their final destinaton. They bud off in a vesicle which will go one of three places:
- Exocytosis – fusion with the cell membrane. Thus the lumenal proteins will be secreted extracellularly, and the membrane proteins will become cell membrane proteins.
- Secretory vesicles – these just stick around as vesicles in the cell until needed – where ‘needed’ means they do eventually undergo exocytosis. In neurons, this is where neurotransmitters are stored until an action potential demands their secretion into the synapse. In the stomach, the cells that produce gastric enzymes keep those enzymes in secretory vesicles until food intake triggers their release into the stomach.
- Lysosomes - where misfolded proteins go to get degraded.
The transport from the trans-Golgi network on to these destinations is different from the other transport discussed above and often involves clathrin (CLT__ genes). Vesicles budding off have a two-layer coat, with adapter protein (AP) complexes as the inner layer and clathrin as the outer layer. The adapter proteins have a target signal with a YXXh motif (h = Φ = any hydrophobic amino acid). Clathrin forms the so-called ‘clathrin-triskelion’ formation shown here:

(Image thanks to Wikimedia Commons user Phoebus87)
Clathrin is also responsible for endocytosis – budding off of vesicles of extracellular stuff (and cell membrane proteins) to come into the cell. This is called clathrin-mediated endocytosis. Receptors in the cell membrane get endocytosed very frequently: the whole population of hormone receptors turns over about every hour, especially when hormones are being received. Taking up the receptor into a vesicle is one way for the cell to cut off the incoming signal until it can be processed.
The plasma membrane notes discuss cystic fibrosis briefly: CFTR is an ABC transporter responsible for pumping Cl- out of the cell (it also lets Na+ in). Loss-of-function mutants don’t pump Cl-, which removes the driving force for osmosis, thickening the mucus and causing breathing problems. There are at least 127 different loss-of-function CFTR mutants (at least, that’s how many Natera tests for) that (if both alleles are disabled) cause cystic fibrosis. The most common mutation is ΔF508, which is ~3% of all European CFTR alleles and about 70% of mutant ones. The loss of that one phenylalanine changes CFTR’s conformation so that the di-acidic exit code (amino acids D565 and D567) that targets CFTR for exocytotic vesicles is no longer correctly exposed and the protein never makes it to the cell membrane [Wang 2004].
discussion section
In section we read Hu 2009, who showed that atlastin proteins are involved in creating the tubular ER network. The evidence came almost entirely from protein-protein interactions. I was surprised this paper was a big deal, because there have been a million papers showing protein-protein interactions for huntingtin, and no one really believes all of them and it hasn’t necessarily gotten us any closer to knowing what huntingtin does or what goes wrong in Huntington’s Disease. But apparently Hu was able to make a pretty clean case for the atlastins’ interactions with reticulons as implying a role in ER formation. It helps that Hu was able to show a ‘genetic interaction’ in addition to a physical (binding) interaction. A ‘genetic interaction’ (I had to look it up) means when “Sometimes mutations in two genes produce a phenotype that is surprising in light of each mutation’s individual effects. This phenomenon, which defines genetic interaction, can reveal functional relationships between genes and pathways.” [Mani 2007].
PrP
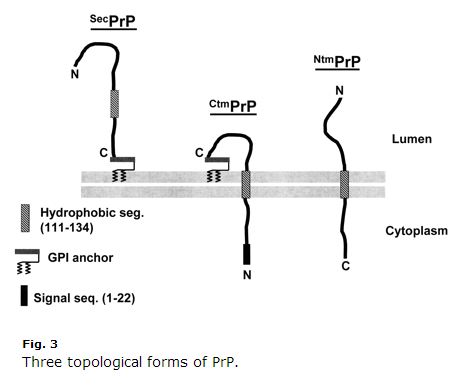
This is a decade old, so some stuff may be outdated, but I found Harris 2003 (ft)’s review of PrP cell biology extremely clear and helpful. Kim & Hegde 2002 was also helpful. PrP is a secretory pathway protein. Its first 22 amino acids (MANLGCWMLVLFVATWSDLGLC) are a signal peptide that causes cotranslational translocation to the ER. Normally, PrP just gets GPI-linked at its C terminus and is anchored to the exoplasmic side of the membrane. But amino acids 111-134 (HMAGAAAAGAVVGGLGGYMLGSAM) are a sort of weak signal anchor sequence (Type II, with the +++ amino acids coming before the signal anchor) that sometimes but not always becomes a transmembrane domain, inverting the C terminus into the lumen. Even more confusingly, that sequence can sometimes just end up as a transmembrane domain without the inversion, so that the N terminus is in the lumen. So there are three membrane topologies of PrP: regular old GPI-anchored, and two transmembrane orientations, as depicted in Harris 2003 Fig 3:
Note how weird CtmPrP is. It’s transmembrane yet also GPI-anchored, and the N-terminal signal peptide is never cleaved off. Normally, the transmembrane forms are < 10% of total PrP. In some laboratory conditions the percentage is higher, and two of the GSS-causing mutations (A117V and P105L) also increase the fraction of CtmPrP to 20-30% of all PrP. Of these three forms, there is a good amount of evidence that CtmPrP is toxic, and that it might play a role in prion formation, though most genetic prion disease mutations (including FFI D178N) do not appear to affect the membrane topology of PrP or the fraction of CtmPrP.
After PrP goes through the Golgi, it is targeted for the cell membrane. But according to Harris, it doesn’t just sit there – it frequently through clathrin-mediated endocytosis and cycles through the cell every ~60 minutes, with some molecules being cleaved on each cycle. Copper stimulates this endocytosis of PrP. Most genetic prion disease mutations change the localization of PrP – usually when a mutation is present, less PrP is found on the cell surface, with more accumulating in the ER.