Protein folding 01: The protein folding problem
These are my notes from week 1 of MIT course 7.88j: Protein Folding and Human Disease, held by Dr. Jonathan King on February 5, 2015.
Introduction
This class focuses on a central unsolved question in biology: how does amino acid sequence direct the folding of proteins? How does cellular machinery assist? We will also discuss human pathologies associated with failures of protein folding. Because the central question here is unanswered, we still do not know what areas of knowledge will prove most relevant to solving it. Dr. King predicts that the problem will be solved - that protein structure will be predictable from sequence alone - within 5-10 years.
The fact that we cannot predict structure from sequence has several important consequences:
- difficulty in achieving native fold when expressing cloned proteins - for instance, recovering natively folded recombinant insulin from E. coli is extremely laborious
- limited progress in de novo design of proteins
- inability to rationally design drugs for protein-folding diseases
In addition to canonical protein deposition diseases such as Alzheimer’s and Parkinson’s, we’ll also consider cataracts, which arise from pathological aggregation of lens proteins. Cataracts are usually not listed as cause of death, despite being responsible for considerable mortality due to accidents associated with reduced visual acuity. One should not think of protein-folding disorders as being diseases of age, even though their incidence is age-dependent. Many environmental insults - say, inhalation of coal dust (black lung) or cigarette smoking - require decades of exposure to cause disease, meaning their incidence too is age-dependent, but age does not cause the disease.
Survey of proteins
One of the smaller, simpler prokaryotes, Methanococcus, has only 1,735 protein-coding genes, with average length 287 amino acids - compare to E. coli, which has 4,285 protein coding genes with average length 317.
How do long protein-coding sequences form over evolution? There are a handful of examples where it has been demonstrated through phylogenetic approaches that two shorter proteins got fused into one. In some cases, two enzymes in the same pathway fused together and ended up doing both jobs.
W and C are the rarest, and L, A and G the most common amino acids in proteins. See this table for frequencies of all amino acids in vertebrates.
A few di- or tri-peptide sequences are rare but very famous - for instance, polyQ, polyFG (in nuclear pore complex proteins), polyP, and G-P-hydroxyproline which is the collagen repeat. But on average, amino acid sequence in proteins appears largely random (or we might say, high information entropy), which has been recognized even from some of the smallest genomes that have been sequenced [Fraser 1995].
Even a 300 amino acid protein has 20300 possible sequences, which is larger than the number of atoms in the universe. This means that the protein folding problem cannot be solved by a lookup table. The set of all sequences that exist in any organism anywhere on earth is but a vanishing subset of possible sequences.
A brief history of empirical protein structure determination
The first growth of protein crystals was achieved by J.D. Bernal and Dorothy Hodgkin (1936-8; see 1964 Nobel Prize in Chemistry). The first 3-D protein structure to be solved was myoglobin, solved via X-ray crystallography by Max Perutz and John Kendrew (1956-8; see Nobel Prize in Chemistry 1962). As of today, Protein Data Bank (PDB) contains 106,293 empirically determined structures (though the number of unique proteins is much smaller).
Until the structure was solved, people expected that myoglobin would possess, at the monomer level, the elegant symmetry of the myoglobin crystal. In fact, its structure is utterly asymmetric. The original structure is available as [PDB# 1MBN]. I show it here in yellow, with the heme in red and sticks shown for the side chains of two histidines coordinating it:
fetch 1mbn
bg_color white
hide everything
show cartoon
color yellow
show sticks, organic
color red, organic
show sticks, resi 64 or resi 97
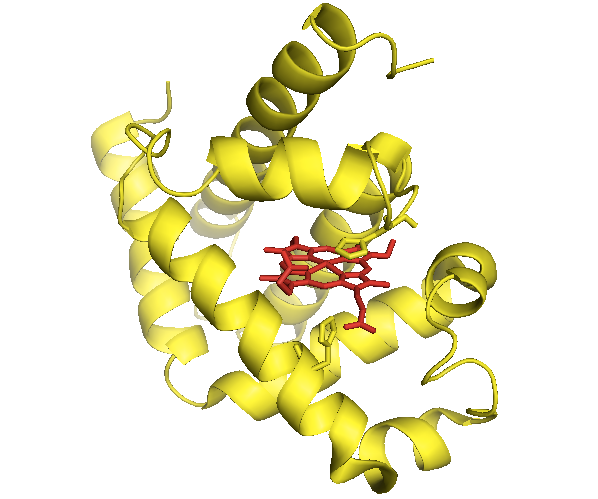
Above: The first protein structure ever solved.
How did proteins acquire such prosaic folds? The structure of DNA was solved >10 years before that of myoglobin was. DNA was helical, and some proteins were already known to be helical, so some people hypothesized a direct templating of DNA conformation onto protein conformation. This theory fell out of favor as experimental data failed to support it - for instance, you couldn’t add DNA to protein and change its conformation. When mRNA was discovered, the theory was revised to posit that mRNA created a conformational template for protein. This too proved experimentally unsupportable. The next theory was that the ribosome physically organizes the nascent polypeptide. This was ruled out quickly, however, as it became clear that ribosomes are all pretty much the same, lacking the diversity of the proteins they produce. Finally, we were left with the “no template” model, which proposed that protein folding proceeded spontaneously. Yet this model remained unpopular for many years, apparently because it didn’t seem very conceptually elegant or exciting.
Eventually, experimental support for the “no template” model arrived in the form of the Anfinsen experiment, the first results of which were published in [Anfinsen 1961]. Anfinsen and colleagues at NIH purified RNAse from the bovine pancreas, an enzyme responsible simply for breaking down the RNA found in dietary grass. Bovine pancreas RNAse comprises 124 amino acids or ~13.7 kDa, and its structure is now known to contain 37% β-sheet, 23% α-helix, as well as coils, turns, and 8 cysteines forming 4 disuflide bonds. Consider the relationship between the number of cysteines and the number of possible permutations of disuflide bonds you could form:
| # of cysteines | # of disulfide bonds | # of combinations |
|---|---|---|
| 4 | 2 | 3 |
| 6 | 3 | 15 |
| 8 | 4 | 105 |
| 10 | 5 | 945 |
So in theory, RNAse could form 105 different disulfide-bonded isomers. But in fact, for RNAse as well as for all disulfide bond-containing proteins now known, only one isomer is actually produced.
Bovine pancrease RNAse was appealing as an experimental substrate for several reasons. It is highly expressed, and available cheaply in large amounts from slaughterhouses. It is extremely stable to protease digestion, a wide range of pH, and a wide range of temperature. It evolved to be super stable, because it operates in the cow gut at pH ~2 and in the presence of high concentrations of trypsin and chymotrypsin. These properties could be leveraged to purify RNAse. Until then, the only proteins that had been appreciably purified were very abundant blood proteins - usually hemoglobin. In fact, Perutz got his myoglobin from whale blood. RNAse had yet another desirable property: it is an enzyme with readily assayable activity. By measuring cleavage of RNA in vitro you could measure how much enzyme function you had recovered.
By this time, chemists already knew that thiol (-SH) groups were the most reactive groups on amino acids. Anfinsen and colleagues therefore expected that the disulfide bonds would prove responsible for the catalytic activity of RNAse. They hypothesized that if they reduced the disulfide bonds with mercaptoethanol, they could abolish RNAse’s activity. You could count the number of reduced (-SH) cysteines using a colorimetric assay based on a small molecule that selectively binds -SH. They were unable to completely reduce all eight cysteines with mercaptoethanol, so they turned to 8M urea. In the process, they discovered that high molarity urea denatures proteins. After they had finally denatured and reduced all four disulfide bonds of RNAse, the catalytic activity was gone. They then dissolved the protein at 1 mg/mL into a phosphate buffer, and then very gradually bubbled in an oxidizing reagent, or alternately dialyzed out the urea and mercaptoethanol. Once the solution turned sufficiently oxidizing, about 15% of the original catalytic activity was recovered. This was taken as evidence that the information required for protein folding was encoded in the polypeptide chain itself, because the protein was able to “remember” and reassume its native conformation.
While this represented an enormous breakthrough, it also led to several misconceptions which took decades to break down. One conclusion that many people drew from the Anfinsen experiment is that the native state of a protein is the lowest free energy state. This attitude may have retarded progress in the field, as it implied that a protein’s search for its native state is simply combinatoric rather than following a specific and dissectable pathway. Cyrus Levinthal eventually calculated the number of possible folds for a protein, and found that the number was astronomical, and there was no way a protein could sample every state. Even after this calculation was published, though, it was years before people accepted the empirical evidence that, indeed, a great many proteins simply do not adopt a native fold spontaneously. Many researchers tried to get actin and tubulin to spontaneously refold in solution, each thinking they were the first to try, then failing, and keeping their unpublishable failure a secret, thus allowing the next fool to do the same. It took many decades of failure before it was discovered that actin and tubulin can fold only with the help of specific chaperones, which will be introduced later this semester. Another, later, realization has been that the native state is not necessarily the lowest energy. In attempts to purify proteins, it was often the case that a large amount of aggregated protein would be discarded in the pellet, and only the properly folded soluble protein would be studied and reported. Calculations regarding free energy and so on were all done on only the soluble fraction, with the rest swept under the rug. It has been a long road to the realization that the amyloid state of many proteins is exceptionally stable.
Indeed, even for RNAse, one of Anfinsen’s students later discovered that re-folding RNAse resulted in the production of plenty of species containing incorrect combinations of cysteines being bonded to each other - these species would pellet out and had been ignored in the original experiment. That student went on to discover disulfide isomerase, an enzyme whose physiological function is to reorganize disulfide bonds into the correct configuration.
