Prion2015 Day 4

Thomas Wisniewski
Dr. Wisniewski’s talk spanned his last decade-plus worth of work towards the development of vaccines against peripherally acquired prion disease in mouse models [Goni 2005, Goni 2008] and more recently in deer [Goni 2015].
He then moved on to reviewing both active and passive immunization approaches for Alzheimer disease, following along the lines of a review he recently published [Wisniewski & Goni 2015]. He listed the different types of toll-like receptors and the opportunities for stimulating innate immunity, and then presented some results from a study last year in which he targeted TLR9 in a mouse model of Alzheimer disease [Scholtzova 2014]. He then discussed some recent unpublished results and plans for future experiments.
Randal Halfmann
Dr. Halfmann began with a review of some interesting complexities of yeast prions. He noted that some prion states result in either cell death, or multicellular cooperative behavior.
He discussed Zhijian Chen’s discovery that the mammalian protein MAVS forms a prion as part of its innate immune signaling role [Hou 2011]. The mechanism is that MAVS polymerization along the mitochonrdiral outer membrane activates NF-ΚB and ultimately interferon. MAVS is unlike other prions. Diverse prion substrates such as PrP, tau, and Sup35 are united by having intrinsically disordered domains, while MAVS is a highly structured alpha-helical protein with a well-characterized crystal structure. Dr. Halfmann wanted more evidence that MAVS is a bona fide prion, so he partnered with Dr. Chen [Cai 2014]. They fused the MAVS prion domain, called CARD, to Sup35’s translation termination domain, and found that it was able to form a prion and inactivate the Sup35 native function, just like the Sup35NM domain can do. The prion state was also transmissible via cytoplasmic mixing (cytoduction), and overall behaved exactly like [PSI+]. They also found that a mammalian protein, RIG-I, involved in the same innate immune pathway, can trigger MAVS prion formation in yeast. And they found that a series of mutants that were already shown to prevent pathway activation [Liu 2013] can prevent prion polymerization in yeast, and can prevent RIG-I from nucleating prion formation. Then they did the converse experiment, which Dr. Halfmann said he thought would never work: they tried to reconstitute the behavior of MAVS in a mammalian cell system using a yeast prion domain. They swapped out the CARD domain of MAVS for the Sup35NM domain. They found that the interferon response could then be activated by inoculating Sup35NM fibrils into the cells. This effect had “like seeds like” specificity - adding PrP fibrils did not induce an interferon response.
Finally, they repeated the entire battery of experiments for a different putative functional mammalian prion, called ASC. The results were very similar to the results for MAVS. For both MAVS and ASC, they found that the prions do not meet the definition of amyloid - they have some morphological similarities but different tinctoral properties, for instance not binding ThT. The structure of these prions has now been solved to 3.8Å by cryo-EM by [Lu 2014]. Here is the structure [PDB# 3J63]:
fetch 3j63
hide everything
bg_color white
show surface
color 0xFF2015

Soon thereafter, two other groups reported that these prion particles are released by cells and taken up into other cells, serially propagating the innate immune response [Franklin 2014 and I didn’t catch the other citation].
He has since explored the larger realm of prions in innate immunity and cell death / apoptosis. He believes there are a lot of prions in these gene families and that many of them are conserved across very distantly related species. For instance, he mentioned a recent report that the mammalian RHIM motif, found in some cell death pathway proteins, is orthologous to Porospoda’s HET-s [Kajava 2014].
Q&A
I didn’t follow exactly all of the arguments, but there was some back-and-forth between Dr. Halfmann, Claudio Soto, Holger Wille and one other person I couldn’t see, about whether ASC and MAVS really meet the criteria to be a prion, and if they do meet it, then how are actin and tubulin not also prions? Dr. Halfmann replied that he thought it was important not to focus on the terminology.
Also, Peter Klöhn asked whether they’ve determined how exactly the aggregates cause cell death - for instance, have they used caspase inhibitors to show that it’s caspase-dependent. Dr. Halfmann replied that it is indeed caspase-dependent.
Tricia Serio
Dr. Serio presented unpublished data on the role of chaperones in the nucleation and curing of prions in yeast.
Jesús Requena
Dr. Requena explained the use of limited proteolysis plus mass spectrometry to map the structure of PrPSc, an approach he applied to anchorless prions a couple of years ago [Vazquez-Fernandez 2012]. As stated in his oral abstract O.14 here, he has now applied this approach to PMCA-derived recombinant PrP prions (provided by Joaquín Castilla), and he sees the same signature, thus suggesting that brain-derived and PMCA-derived prions share a common structure. He also spent a few minutes discussing his poster #P.112 here, which argues that the protease accessibility sites in multiple PrPSc strains, together with other lines of evidence, suggest a β-solenoid structure.
Q&A
Q. Jiri Safar: There is a risk that your preparations contain a mixture of infectious prions and non-infectious amyloid species, and that the species you are measuring is not the infectious one. Have you considered looking at known non-infectious recombinant amyloid as a negative control?
A. Jesús Requena: A problem is that to some degree, the data from non-infectious amyloids suggest a similar secondary structure - the same portions of PrP form beta strands - the question is whether the larger superstructure is a solenoid or something different. But yes, you’re right that there could be some degree of mixture.
Anton Gorikovskiy
Dr. Gorikovskiy explained in greater detail his and Reed Wickner’s published work on locating the folds in the parallel in-register intermolecular β-sheet (PIRIBS) structure of Sup35 amyloid [Gorkovskiy 2014], which Dr. Wickner discussed briefly in his keynote. He explained the utility of dipolar recoupling as a method to discriminate between scenarios of parallel in-register vs. anti-parallel vs. parallel out-of-register structures. This method lets you measure the distance between copies of the same residue in different molecules of the protein. Based on this distance you can tell which residues are or are not parallel and in-register. They applied this method to a variety of Sup35 mutants with 13C-labeled isoleucine introduced at various points. He also explained how they took great care to ensure that the species of amyloid they were measuring in these mutants were able to propagate in vivo. They identified specific residues that appear to be parallel in-register and other residues that are clearly not parallel in-register, which therefore appear to represent the folds of the PIRIBS structure (loops between beta sheets).
Q&A
Mark Zabel announced that it is Dr. Gorikovskiy’s birthday and that his brother sponsored cake and ice cream which will be served momentarily.
Eric Ross
Dr. Ross studies the amino acid composition and sequence requirements for prion formation. He began by discussing his lab’s in silico studies [Toombs 2010, esp. Figure 6], indicating that the proteins that form prions in yeast are those that have both a high “prion propensity” and a low “order propensity”. A series of experiments from the Ross lab have explored the requirements for prion activity, and the ways in which different amino acids confer differing amounts of “prion propensity”, but there are also poorly understood sequence context effects [Toombs 2011, Gonzalez Nelson 2014, MacLea 2015]. As a result, there has been some success in bioinformatic prediction of prion domains [e.g. Ross 2013] but there is much work left to do.
He concluded with some speculation on how evolution both creates prion domains, and keeps them in check. He argued that genetic drift will tend to push potential prion domains right to the edge of aggregation, to the point where a single additional mutation may be sufficient to cause prion formation. He also noted that once a prion domain is formed, its aggregation propensity can be greatly increased by the duplicaton of multiple repeating copies of the domain.
Gerold Schmitt-Ulms
Back when he worked in the Prusiner lab, Dr. Schmitt-Ulms discovered that neuronal cell adhesion molecule 1 (NCAM1) is the major protein that physically interacts with PrPC [Schmitt-Ulms 2001]. They used formaldehyde to cross-link PrP to any interacting factors, ran it on a gel to separate cross-linked PrP from PrP monomers, and then boiled the cross-linked bands to separate the interactors. They found abundant NCAM1. At the time, the Prusiner lab’s interest was in identifying Protein X, so when they found that NCAM1 knockout mice had unaltered prion disease incubation times, they lost all interest in the protein.
Years later, once he had his own lab, Dr. Schmitt-Ulms rekindled his interest in NCAM1. This was inspired in part by the connection between PrP and ZIP proteins. He discovered that PrP and the ZIP proteins are paralogous - indeed, ZIP10 has about as much similarity to PrP as Dpl does [Schmitt-Ulms 2009]. Also, zebrafish studies identified similar deficits in epithelial-mesenchymal transition in embryos with zebrafish PrP1 or ZIP1 knocked down by morpholino [Yamashita 2004, Malaga-Trillo 2009].
Most of the talk consisted of unpublished data from experiments by his student Mohad Mehrabian. These data are summarized in his abstract O.15 here.
He ended by saying how excited he is that Joel Watts has joined his department at University of Toronto, and that he looks forward to many future collaborations.
Lindsay Parrie
Dr. Parrie is a postdoc in Richard Bessen’s lab, and if you’ve followed Dr. Bessen’s work you know he has a longstanding interest in the role of olfactory neurons and olfactory mucosa in the uptake, neuroinvasion, and shedding of prions [Bessen 2009, Bessen 2010, Bessen 2012, Crowell 2015]. Dr. Parrie presented unpublished data (see abstract O.16 here) on the effects of prion infection on olfactory sensory neuron proliferation after injury.
Eri Saijo
Dr. Saijo reviewed the Horiuchi lab’s development of the 6H10 antibody. It was created by immunizing PrP knockout mice with purified Obihiro strain PrPSc, and the antibody exhibits conformational selectivity for PrPSc over denatured PrPSc [Horiuchi 2009]. As summarized in her abstract, O.12 here, she presented unpublished data indicating that this antibody also exhibits differential binding between PrPSc of different strains, and is able to discriminate between RML and 22L prions.
Q&A
Q. Have you thought about testing the 15B3 antibody?
A. No.
Holger Wille
Dr. Wille started from his early X-ray fiber diffraction experiments in the Prusiner lab [Wille 2009], which showed “meridional reflections” at 4.8, 6.4 and 9.6Å. He now interprets these data to indicate a 4-rung β-solenoid structure where each PrP molecule occupies 4 rungs and spans a total of 19.2Å along the long axis of the fibril. He then presented some unpublished work applying the same method to a variety of different prion strains. He also noted that non-infectious in vitro amyloids give a different structure [Wan 2015].
While they were doing this work, the high-resolution structure of the HET-s prion was solved, revealing it to be a 2-rung β-solenoid [Wasmer 2008]. They therefore prepared the fibrils and saw 4.8 and 9.6Å signals virtually identical to the ones they saw for PrPSc, except for the lack of a 6.4Å signal, reflecting a 2-rung structure for HET-s compared to a 4-rung structure for PrPSc [Wan 2012].
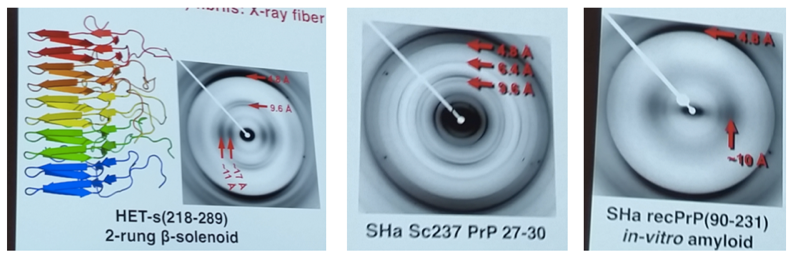
Left: the known β-solenoid structure of the Porospoda HET-s prion, and its X-ray fiber diffraction pattern. Center: the X-ray fiber diffraction pattern of brain-derived PrPSc, which is similar to that of HET-s but consistent with 4 rungs rather than 2. Right: the X-ray fiber diffraction pattern of a non-infectious, in vitro-derived PrP amyloid, which is distinct from the other two.
His talk then moved from fiber diffraction to cyro-electron microscopy. He said that EM is currently undergoing a revolution because direct electron detectors are making it possible to collect much higher-resolution images. He presented unpublished data from 3-D reconstruction and single particle analysis.
Q&A
Q. Detlev Riesner: One turn of the β-solenoid has three bending points. Does your model predict where these are?
A. The left-handed has three bending points, but the right-handed is less well defined and may not be as regular, so I stepping away from this a bit. But Jesus Requena’s data presented earlier today based on limited proteolysis do provide some insight as to where the bends might be.
Q. Byron Caughey: Impressive stuff, I have to think about it. Have you looked at anchorless fibrils with cryo-EM?
A. No, so far we’ve only looked at RML with cryo-EM, and we are preparing to do it with CWD and BSE.
Jiri Safar
Dr. Safar presented recently published work from his lab and Witold Surewicz’s lab on the structural determinants of phenotypic diversity across prion strains [Safar 2015]. He started from clinical data, noting that mean survival time after disease onset is almost four times longer in MM2 sCJD than in MM1 sCJD. His earlier data from conformation-dependent immunoassay, QuIC, and PMCA indicated that it is the more unstable prion strains that replicate faster and exhibit more rapid disease progression in vivo [Kim 2012]. On the face of it, those data would seem to be in agreement with the model Jonathan Weissman has proposed for yeast prions [Tanaka 2006, Toyama 2007]. However, his latest data no longer support that model. He and Dr. Surewicz then performed hydrogen/deuterium exchange on MM1 sCJD vs. MM2 sCJD prions and found large differences in solvent accessibility, with MM2 more accessible towards the central region (~117 - 144) while MM1 is more accessible in the C terminus (~153 - 224) [Safar 2015 - Figure 3]. This indicates a difference in backbone conformation. The H/D exchange difference is particularly great for histidines, indicative of “quaternary packing arrangements”. The two strains also differ in particle size and replication rate in PMCA and QuIC [Safar 2015]. The overall interpretation is that the kinetics of replication are ultimately driven by an interaction between two factors: 1) PrPSc stability and 2) the affinity of PrPSc for PrPC.
Q&A
Q. Michael Geschwind: How does the replication rate of MM1 versus MM2 relate to the faster clinical course?
A. This is exactly what is surprising. Jonathan Weissman’s model indicates that more unstable strains replicate more quickly, while we find that MM1 is more stable but replicates more rapidly.
Jason Bartz
Dr. Bartz began with his early work showing that differences in incubation time for prion strains represent an interaction between strain and inoculation route [Bartz 2005]. In hamsters, the DY TME strain not only has longer incubation times than HY on i.c. inoculation, it also is unable to neuroinvade after oral or peripheral inoculation. In fact, it appears to not even replicate in lymphoid tissues, as the hamsters inoculated peripherally with DY had no evidence of PrPSc in their spleen or lymph nodes after 60 or 120 days [Bartz 2005]. I quickly got lost in the details, but he discussed several experiments on the transport proprties of different strains, for example their ability to cross epithelial layers after extranasal inoculation [Kincaid 2012]. He also discussed new results from his lab showing the presence of RT-QuIC seeding activity in blood throughout the incubation period [Elder 2015]. The summary conclusion of all of this work was that DY’s inability to neuroinvade is due to an inability to colonize lymphoid organs. Then he presented further work dissecting the mechanism, some of which was unpublished.
Edward Pokrishevsky
Dr. Pokrishevsky presented his work in Neil Cashman’s lab suggesting that incorrect localization of FUS and/or TDP43 in the cytosol can induce the prionization of wild-type SOD1 [Pokrishevsky 2012]. These findings offer a possible link between TARDBP or FUS-linked ALS and SOD1 ALS, and a possible common disease mechanism between genetically disparate but clinically similar forms of ALS.
Closing ceremonies
Glenn Telling thanked everyone. He also praised the attendees for having made extraordinary progress in prion research in the last 20 years, and for being so collaborative. He expressed his hope that prion researchers will continue to be an outsanding example of being collegial and collaborative and attracting young talent. Mark Zabel announced the poster prize winners: Malin Reiten, Yo Ching Cheng, Sean Cascarina, Tim Kurt, and Cheng Fang. Hilariously, one of the prizes was David Borchelt’s brewery tour ticket. Hidehiro Mizusawa announced that Prion2016 will take place at Hitotsubashi Hall, National Center of Sciences Building, Tokyo, Japan on May 11-13, 2016 (Wed-Fri). Before the main meeting there will be a teaching course May 8-10, and workshops the morning of May 11.


The venue is the building highlighted in purple in the second image. Behind it is the Emperor’s Palace.
