Screen reveals antiprion compounds that act indirectly
 Read with caution! This post was written during early stages of trying to understand a complex scientific problem, and we didn't get everything right. The original author no longer endorses the content of this post. It is being left online for historical reasons, but read at your own risk. |
Poncet-Montagne 2011, of UCSF, brings us the results of a 2,160-compound screen for antiprion molecules with a (perhaps) surprising conclusion: all the molecules found to exhibit antiprion properties do so through third parties. None interact directly with PrP.
The methods for this screen are described in quite a bit of detail, which I took as a chance to learn how a high-throughput assay for prion therapeutics can be designed. The screen, which included “known drugs, bioactives, and natural products” (presumably MSDI’s Spectrum Collection), was conducted at the Small Molecule Discovery Center, UCSF’s high-throughput screening facility. The initial screen used mouse neuroblastoma cells (N2a cells) infected with RML prions (this combination is called ScN2a cells) in 96-well plates, coincubated with candidate molecules for 5 days. The negative control for each compound was ScN2a cells treated with DMSO, in which the tested compounds had been solubilized; the positive control was ScN2a cells treated with quinacrine. The overall screening methodology is called ELISA and had been described for ScN2a cells in an earlier screen done by the same lab [Ghaemmaghami 2010]; the full workflow for the assay is in Fig 1A from that 2010 study. So it took some digging, but basically the readout from this assay is as follows:
For each compound, cytotoxicity was measured by a calcein-AM assay, and the effect on PrPSc levels was detected by ELISA following PK digestion and PTA precipitation.
So basically, the ScN2a cells were coincubated with the compounds for 5 days, then subjected to proteinase K, which should digest PrPC but not PrPSc, and then immunoblotted with anti-PrP antibodies to get a visual readout as to whether PrP was still present. The images were analyzed using ImageJ (they don’t say exactly how, but here’s an example of using ImageJ on Western blots). So that’s the readout: the less PrP is still present after proteinase K digestion, the less PrPSc there must have been, and so the better the compound’s antiprion properties. So the outcomes of this screen are only translatable into therapeutics to the extent that proteinase K resistance correlates with infectivity and toxicity. I understand that this correlation is fairly strong but not absolute– I’m still looking for a citation that discusses the issue in more detail.
As an aside, I found it curious that the screen used quinacrine as a positive control. This implies that quinacrine’s antiprion properties are so well-established that they are actually the standard by which to judge other compounds. I don’t know how to square this with the fact that Barret 2003 showed that quinacrine’s antiprion activity was less than that of tetracyclines. It is also peculiar that quinacrine works so well in vitro yet ultimately failed clinical trials [Collinge 2009]. Hopefully more on that in a future post.
Now, what I just described above was just the initial discovery assay as best I understand it. A “hit” was defined as “a compound that reduced PrPSc load by 40% with less than 20% cytotoxicity” — i.e. it both works and doesn’t kill the cells. The assay of 2,160 compounds yielded 206 hits, which largely overlapped with the hits from another screen by Kociscko 2003. 206 was too many to follow up on, so the authors randomly chose 40 of the 206 compounds.
These 40 compounds then went through the same ELISA screening protocol again and 16 of them passed it again.
So then came the question, what exactly are these 16 compounds doing? The authors had a whole battery of tests to try to assess this. Here is an incomplete list:
- Isothermal titration calorimetry to figure out if the compounds were interacting directly with PrP. Answer: no. None of them interacted directly. The positive control was suramin, which does interact directly with PrP (in a bad way, inducing aggregation). This part of the study used synthetic peptides of mouse PrP 89-230 rather than full-length PrP. Not sure why.
- Circular dichroism to measure whether the compounds affected the melting point of MoPrP(89-230). If they did, that would suggest they were binding to PrP. Answer: no, they didn’t bind to PrP. The positive control was TMAO, which does bind to and stabilize PrP.
- Differential scanning fluorimetry (DSF) to measure whether compounds interact directly with PrP. After the above two experiements, the authors wondered if the fact that the 16 hits didn’t interact with PrP was because none of the original 2,160 compounds did so, or because interacting with PrP is a poor predictor of antiprion activity. So they actually went back and screened all of the original compounds with DSF. Answer: none of the original compounds interact with PrP (again, this is actually MoPrP(89-230), not full-length PrP). The authors did not find this surprising, because:
NMR and crystallographic studies indicate that the three-dimensional structure of PrP lacks deep cavities normally required for high-affinity binding interactions with small molecules, and as such, few molecules within an unbiased diversity chemical library are expected to bind PrP
- The authors purified PrPSc from mouse brains and tested by Western blot whether any of the 16 hits increased the proteinase K sensitivity of this PrPSc in this purified chemical setting. Answer: no. They don’t. Interesting. So when you add these compounds to ScN2a cells, digest with proteinase K and then Western blot, you find that there is less PrPSc, yet when you do the same experiment with purified PrP and no cell present, the effect goes away. So if the compounds do increase the proteinase K sensitivity of PrPSc in cells, they must do so via some indirect pathway dependent upon cellular process or other proteins or small molecules present in the cell.
- Screening of the 16 hits on uninfected N2a cells followed by Western blotting (no proteinase K step here) to simply see if the compounds reduce the amount of PrPC present in the cells. Answer: yes, a few of them do. It makes perfect sense that reducing the overall amount of PrP would be one route to antiprion activity. There’s plenty of evidence out there that PrP infectivity is concentration-dependent. For instance, Jackson 2009 was able to achieve FFI prion infectivity only in mice overexpressing wild-type PrP and not in mice expressing wild-type PrP at normal levels. Jeffrey W. Kelly also likes to talk about how “protein aggregation is a concentration-dependent process”. So if a compound simply reduced the total amount of PrP present, that could in itself have therapeutic value even if the compound didn’t target PrPSc with any specificity. That looks to be at least part of the story for some of these 16 compounds.
- A flotation assay to see if any of the 16 compounds disrupt lipid rafts. The authors offer that “an alternative mechanism for decreasing PrPSc formation is to alter the integrity of lipid rafts, membrane microdomains where PrPC is known to localize”, which has been the rationale for some of the studies on statins and prion disease, though those studies ultimately concluded that cholesterol was not the statins’ mechanism of action on prions. Anyway, two of the 16 hits in this study were statins. Result: of the 16, only lovastatin appeared to disrupt lipid rafts. However, several of the compounds appeared to affect PrP localization.
- Two more experiments on amcinonide, which was the compound which reduced the concentration of PrPC the most in the above experiment on the uninfected N2a cells. The authors wanted to know at what step amcinonide acted on PrPC levels: by reducing transcription? Nope, qPCR showed no effect on PrP mRNA levels. So then the authors stopped translation of PrP with cyclohexamide, and then added amcinonide to see how it affected the degradation of the already-translated PrP. The half life of PrP was reduced from the 12-18h range down to 6h. So amcinonide acts by increasing PrP degradation.
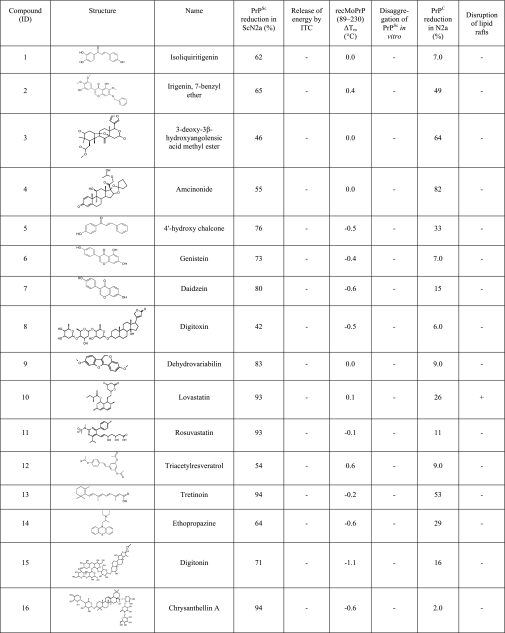
So those were the experiments. Here are the 16 molecules:
The authors haven’t yet reported any in vivo validation of these 16 hits, so it’s on the early side to get hopeful that any of these compounds could be used as prion therapeutics. But what’s really interesting from this screen is the finding that maybe direct interaction with PrP is not the route to drug development. In fact, the authors even question the idea that any compounds previously identified by other studies really directly interact with PrP in a way that is specific, reproducible, and highly affine. They think this idea of direct interaction with PrP is a dead end and that assays focused on direct interaction hold little promise:
The results are consistent with the reported crystal and NMR structures of PrPC, which indicate the lack of prominent structural clefts capable of ligand binding (32–35). Various compounds have been reported to bind PrPC in vitro. However, these interactions have been shown to be nonspecific (e.g. anionic tetrapyrroles (48)), irreproducible in alternative binding assays (e.g. Gn8 (10)), or have millimolar affinities (e.g. quinacrine (49)). More recently, it was reported that a cationic tetrapyrrole (Fe(III)-TMPyP) can specifically interact with the folded domain of human PrP with micromolar affinities, although the large size of the compound may preclude it as an effective brain-permeable clinical therapeutic (50). The results of our study suggest that a drug-discovery screen focused on the identification of specific PrP-interacting molecules, e.g. protein X, is unlikely to identify many efficacious compounds.
Instead, antiprion compounds are likely to act through a third party molecule or “protein X”:
In protein aggregation diseases, including prion diseases, a common target for pharmacological intervention has been the aggregating molecule itself (46, 47). Although we were able to identify 16 nontoxic compounds that were active against prion formation in cell culture, none of the 16 molecules interacted with PrPC or PrPSc in vitro, as evaluated by various biophysical techniques. These observations imply that at least one other cellular target (protein X), likely several given the diversity of the active molecules, modulates PrP conversion within the cell.
This idea of a “protein X” which is involved in prion formation interesting. If true, then genetic variation in the gene coding for such a “protein X” should be a significant modifier of prion disease. For instance, homozygous loss of function mutations in protein X might render someone resistant to genetic prion disease. Or variants which increase the affinity of protein X for interaction with PrPSc might be risk factors for sporadic prion diseases. Yet as mentioned in the GWAS post, genome-wide association studies have yet to identify any risk factors for prion disease other than, of course, loci within PRNP itself.
This also leads me to the question, would “protein X” be the same for different prion diseases? Perhaps different proteins would have different affinities for different misfolded conformations of PrP, or perhaps the different strains have different levels of dependence on things like lipid rafts which protein X could be involved in indirectly. CJD, FFI, and GSS each have different patterns of pathology across the various brain regions, different signatures in terms of plaque formation and vacuole formation, and so on. Roles for multiple different “X” proteins could be one explanation. If so, then antiprion therapeutics might not always translate well across prion strains, but
In concluding, the authors identify a couple of mechanisms for potential antiprion therapies based on their results. They remind us that PrP knockout mice are resistant to prion disease and point out that compounds like amcinonide which increase degradation of PrP could have therapeutic value. They also discuss the two statins — lovastatin (Mevacor) and rosuvastatin (Crestor) — in their hit list and the potential role of lipid rafts as necessary for prion formation. This notion of statins disrupting lipid rafts and therefore prion formation is difficult to reconcile with the fact that the four studies on statins I reviewed last week generally seemed to conclude that the mechanism of action of statins on prion disease did not involve cholesterol and, moreover, that mice treated with statins actually had more PrPSc than untreated mice at the time of death. update 2013-05-24: it’s actually more complicated and hard to draw conclusions about mechanism of action from those studies, see this new post. For what it’s worth, these are different statins than tested in those studies. I dug up the compound list for MSDI’s Spectrum Collection, which has since grown to 2,320 compounds, and it does include both simvastatin and pravastatin. These are old enough drugs that I am assuming they were also in the 2,160 initially screened in this study. I’d be curious whether they were some of the 206 initial hits that just didn’t get randomly selected for followup.
As for the other 13 compounds, they state that we really don’t know the mechanism of action, other than it’s probably not direct interaction with PrP.
All in all, the 16 hits validated here are a pretty interesting mixture of FDA-approved drugs, herbal compounds, and experimental compounds:
- Isoliquiritigenin. This is one of the ‘natural’ compounds, a licorice phenol; under “source”, MSDI simply says “widespread in Fabaceae“. It acts on sirtuins, those magical proteins which some people think are the key to immortality. (Sirtuins are also the target of resveratrol, that hyped-up compound in red wine).
- Irigenin, 7-benzyl ester. A derivative of another natural compound from iris rhizomes.
- 3-deoxy-3-beta hydroxyangolensic methyl ester. No idea what this compound’s deal is.
- Amcinonide. This is actually an FDA-approved drug, though for topical use only. It’s one of those corticosteroids they prescribe as a cream for itchy skin. Branded as cyclocort.
- 4′-hydroxy chalcone. No idea.
- Genistein, another herbal compound, an isoflavone present in a lot of legumes as well as coffee. Despite the ‘herbal’ status, there seem to be a lot of references to molecular interactions that have been demonstrated for genistein. Among them, Singletary 2008 cites evidence that it promotes autophagy and apoptosis in cancer (good and bad from the perspective of prion disease??)
- Daidezin, another isoflavone present in soybeans among other things. It’s a legal dietary supplement in the U.S. but not considered a drug.
- Digitoxin, true to the ‘toxin’ in its name, was once the murder weapon (it’s present in foxglove) in an Agatha Christie novel. But in lower quantities it’s also a drug. It interferes with sodium-potassium pumps in cardiac muscle, thus increasing intracellular calcium concentration in the ER, so that when the cell is stimulated it releases more calcium, resulting in a stronger contraction. It was used to against heart failure for a while, though it seems to have fallen from favor.
- Dehydrovariabilin. No idea.
- Lovastatin (Mevacor), a statin.
- Rosuvastatin (Crestor), another statin.
- Triacetyl resveratrol. According to the link at left from Enzo Life Sciences, it’s regular old resveratrol plus a phenol group to increase its half life in the body. As mentioned above, resveratrol is a sirtuin activator and much-hyped anti-aging compound.
- Tretinoin, the acid form of vitamin A, in use as a drug for acne and cancer.
- Ethopropazine, aka profenamine. A drug prescribed to control the symptoms of Parkinson’s disease. It antagonizes muscarinic ACh receptors and so controls the involuntary movement in Parkinson’s.
- Digitonin, which is only one letter different from digitoxin simply because they both come from foxglove (Digitalis purpurea). It water-solubilizes lipids.
- Chrysanthellin A. Couldn’t find much about this online, and it’s actually not in MSDI’s spreadsheet, at least under that name. This material safety data sheet from a supplier in Mumbai suggests it is probably not ready for prime time as a drug.
Again, it’s an interesting list but remember these were merely the hits from an in vitro screen with no in vivo validation yet. This blog does not provide opinions on the efficacy of any medical treatment and nothing said here should be construed as medical advice.