Proteolysis of prion protein in the central region: alpha and beta cleavage
Proteo- means protein, and -lysis means clevage. This post is about where and how prion protein (PrP) is cleaved in its central, highly conserved region. There are two types of PrP cleavage in this region, known as alpha and beta cleavage.
the central region of PrP
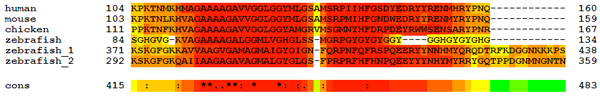
If you zoom out from Mammalia and look at other vertebrates, there is only one large part of PrP that is really conserved even between us, chickens and zebrafish. You can see it in this multiple alignment from this post:
This stretch runs from human amino acids 104 – 160, with particularly strong conservation at 112 – 150 or so.
The first half of this tract, approximately 112 – 134, is the so-called “hydrophobic core”, rich in A, M, L and V that gets embedded in the membrane in CtmPrP and NtmPrP [reviewed in Harris 2003 (ft)]. Mutations that increase the hydrophobicity of this stretch cause GSS and tend to increase the fraction of PrP that ends up with a CtmPrP transmembrane topology [Hegde 1998 (ft)].
Overlapping with the “hydrophobic core” are codons 106-126, believed by some to be the toxic region of PrP. When it is synthesized as a standalone peptide, PrP 106-126 forms amyloid fibrils [Tagliavini 1993 (ft)], and is neurotoxic to cultured hippocampal cells [Forloni 1993], but only if those cells express PrP [Brown 1994]. Conversely, ΔCR PrP, which precisely lacks this “Central Region”, is highly toxic and neonate lethal, though its toxicity can be reduced by co-expression of wild-type PrP [Li 2007]. All this is quite interesting and certainly, together with the phylogenetic conservation of this region, says something important is going on there.
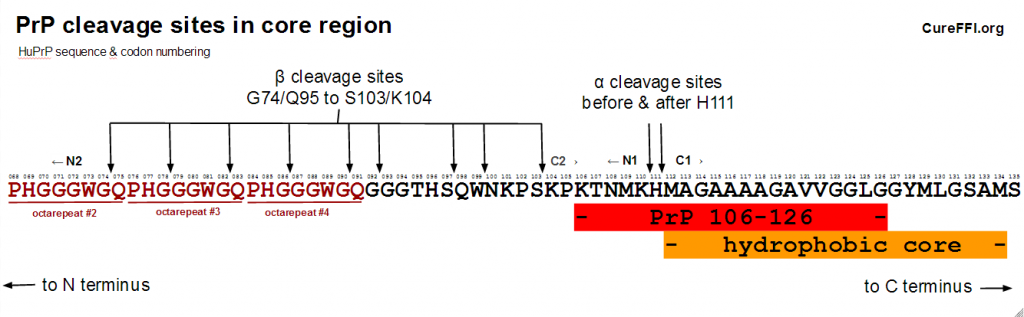
This central region of PrP – between the octapeptide repeats (codons 60-92) and the more structured C-terminal region (beginning with beta sheet 1 at codon ???) – also turns out to be where most of the action is in terms of proteolysis. PrP can undergo so-called “alpha cleavage” before or after codon 111 to yield two fragments dubbed N1 and C1, or it can undergo “beta cleavage” which occurs before codon 91 or earlier, yielding fragments called N2 and C2. Here’s a diagram:
alpha cleavage
Cleavage before or after H111 to generate the N1 and C1 fragments, now called alpha cleavage, was first reported in chicken PrP [Harris 1993 (ft)] and later confirmed in human samples [Chen 1995 (ft)]. Some of the proteolytic enzymes involved in this cleavage were identified (in human cells) several years later [Vincent 2001 (ft)]. The best introduction I found to the subject is a recent review [Liang & Kong 2012].
The ADAM family of proteolytic enzymes – originally from one enzyme dubbed “a disintegrin and metalloproteinase” – are involved in alpha cleavage of PrP. The original report [Vincent 2001 (ft)] implicated ADAM10 and ADAM17 (latter formerly known as TACE). ADAM9 [Cisse 2005 (ft)] and, in muscle at least, ADAM8 [Liang 2012] have also been added to the list. There’s still a fair bit of controversy over which of these is directly involved and how they are regulated [Liang & Kong 2012]. The cleavage event probably does not occur on the cell surface. Instead it is thought to occur either in endocytic vesicles [Shyng 1993 (ft)] or the late secretory pathway [Walmsley 2009].
Alpha cleavage appears to occur quite often in uninfected brains and there is about half as much C1 present as there is full-length PrP [Chen 1995 (ft)].
beta cleavage
As shown in the diagram, beta cleavage can occur at a variety of sites. Wherever it occurs, it is said to generate fragments called N2 and C2. A bit of history is in order to clarify the distinction between C2 and PrP 27-30.
The first protocol for homogeneous purification of PrP used proteinase K digestion as one step [Prusiner 1983]. This protocol applied to infected hamster brains yielded a 27 – 30 kDa protein dubbed PrP 27-30, so pure that it was possible to read the N-terminus of the PK-resistant protein fragment, which started with GQGGGTH [Prusiner 1984], i.e. starting with the equivalent of human codon 90. But when polyclonal antibodies were finally raised against PrP [Bendheim 1984], it was discovered that they not only pulled down PrP 27-30 from infected hamster brains, but also pulled down a 33-35 kDa version of PrP in both infected and uninfected hamster brains [Oesch 1985]. Proteinase K was capable of completely digesting PrP 33-35 from uninfected brains, but only digested part of PrP 33-35 from infected brains, resulting in PrP 27-30.
The PrP 27-30 pulled down by the antibodies without PK digestion probably represented C2 fragments that had already been cleaved in the brain, while PrP 33-35 was full-length PrP, which in infected brains contained a protease-resistant fragment (approximately? exactly?) coincident with C2.
We now know that the protease-resistant fragment varies in size in different prion strains [Parchi 1997]: so-called “type 1″ PrPSc contains a 21 kDa fragment starting at G82, and “type 2″ PrPSc contains a 19kDa fragment starting at S97 [reviewed in Parchi 2011]. (Those are the molecular weights after deglycosylation.)
We also know now that C2 is created even in uninfected brains, just at levels too low for Oesch to have detected. In retrospect, the earliest report of beta cleavage of PrPC may have been [Pan 1992 (ft)]. Pan reported on purification of PrPC from the hamster brain and found “PrPC-I” (full-length PrPC), “PrPC-II” (the C1 fragment), and also reported that:
Interestingly, an immunoreactive 30-kDa band, located just below PrPC-I and often obscured by its intense staining, copurified with PrPC throughout the purification… αP1 raised against a synthetic peptide homologous to the N-terminus of PrP 27-30, also reacted with this 30-kDa protein…
This 30 kDa protein must have been the (diglycosylated) C2 fragment. It contained more of the N-terminus of PrP than the C1 fragment did, so it reacted with antibodies raised against the protease resistant PrP 27-30. It was present in uninfected brains, meaning it was the result of a normal biological cleavage process and suggesting it didn’t necessarily result from protease resistance.
Soon it was confirmed that this C2 fragment is naturally present in uninfected human brains too [Chen 1995 (ft), Jimenez-Huete 1998], though it wasn’t abundant. Chen found there was only about 5% as much C2 as there was full-length PrPC, or about 10% as much C2 as C1, indicating there isn’t a ton of beta cleavage going on under normal healthy circumstances. By contrast, C2 is enriched in prion disease: the brains of CJD patients contain large amounts of C2 – i.e. without proteinase K digestion, one finds both full-length PrPSc as well as PrPSc corresponding to only the C2 portion of the protein, just as in Oesch’s hamsters.
Later on, detection of the N2 fragment confirmed that C2 resulted from cleavage rather than digestion [Mange 2004]. It was also discovered that beta cleavage can be stimulated by reactive oxygen species (ROS) [McMahon 2001 (ft)]. McMahon added hydrogen peroxide (H202 - a source of ROS) and superoxide (O2- - a ROS itself), to cell cultures (CHO cells) and found that incubation of cells with these oxidizing agents caused a loss of full-length PrPC in favor of the C2 fragment. The effect was deemed “probably a non-enzymatic event” – i.e. the ROS don’t recruit a proteolytic enzyme to cleave PrP, but rather, the ROS directly react with PrP to cleave a peptide bond, something which apparently is possible (McMahon cites Kim 1985 (ft)).
It has since been reported that beta cleavage can also be accomplished enzymatically, by calpains [Yadavelli 2004 (ft)] and cathepsin [Dron 2010]. There’s slightly more coverage of this issue in another review [Altmeppen 2012]. The enzymatic beta cleavage has only been reported for PrPSc, not PrPC.
In vivo, PrPSc beta cleavage can occur at G74, G78, G82, G86, G90, G92, S97, W99, or S103 [Parchi 2000, Table 1], with the distribution for type 1 PrPSc centered on G82 (in the third octarepeat) and for type 2 PrPSc centered on S97. These sites are shown in the diagram of the central region above. In vitro, at least, beta cleavage of PrPC has also been reported as early as the second octarepeat [McMahon 2001 (ft)].
disease implications of alpha and beta cleavage
Alpha cleavage cleaves through the core region of amino acids 106-126. This is thought to render the remaining C1 fragment incapable of converting to PrPSc. The best line of evidence for this is that mice constitutively expressing only C1 cannot be infected with prions [Westergard 2011]. Indeed C1 appears to be dominant negative, since Westergard found that its co-expression with full-length PrP lengthens incubation times compared to when only full-length PrP is expressed. It has been reported that N1 is also neuroprotective [Guillot-Sestier 2009, Guillot-Sestier 2012]. While a few other studies have suggested that C1 makes cells or animals more sensitive to pro-apoptotic stimuli (which is bad), Westergard observed no neurological symptoms in C1 mice even with ~7x expression levels. Overall, evidence seems to point to alpha cleavage being a good thing from the standpoint of prion disease.
Beta cleavage, by contrast, leaves PrP 106-126 intact, and when a C2 fragment starting at amino acid 73 is expressed alone, it is still capable of supporting prion disease [Fischer 1996 (ft)]. This, together with the observation that C2 is more abundant in prion-diseased brains than healthy ones [Chen 1995 (ft)], casts beta cleavage as a bad thing from the standpoint of prion disease.
All this is too simplistic – it’s likely that both types of cleavage have a native function, and there is not yet quite enough evidence to say that beta cleavage has any causal relationship with prion pathology or disease progression. But it does appear that the two are mutually exclusive, and that alpha cleavage precludes a PrP molecule from becoming PrPSc.
Interesting aside: all of this, including the nomenclature, closely parallels the cleavage of APP and its relation to Alzheimer’s disease. Amyloid precursor protein (APP) can be cleaved by alpha secretase or beta secretase (BACE1). These are mutually exclusive, and the latter generates amyloid beta (Aβ), so alpha secretase cleavage of APP may be seen as benevolent and preventive. BACE1 inhibitors are under investigation as potential anti-Alzheimer’s drugs. ADAM peptidases are involved in the alpha cleavage of both APP and PrP (probably the reason for the common “alpha” label), and we now know PrP is a receptor for Aβ.
preview
Besides alpha and beta cleavage, PrP can also be cleaved at the extreme C terminus, “shedding” the almost-full-length protein from the cell surface. Shedding will be the subject of my next post.