Descriptive statistics and quality control on variants
I have previously described how to aggregate data from FastQC to check the quality of raw reads from next-generation sequencing, and my GATK exome sequencing pipeline covers how to use bedtools to calculate coverage after alignment. After you’ve completed the pipeline and called variants, you still need to see if the variants you’ve called look reasonable. This post will introduce the descriptive statistics generally used to assess the quality of variants called from exome data.
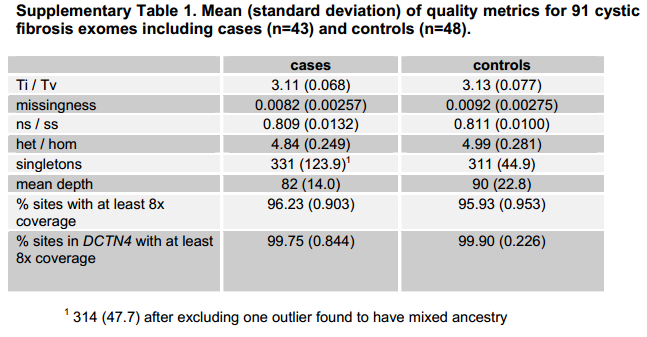
Emond’s cystic fibrosis exome paper specifically calls out the following metrics for that study’s exome variants in the supplement:
What are all of these metrics? I had to look most of them up. Here are some definitions (as best I understand):
- Ti/Tv (sometimes called Ts/Tv): the ratio of transitions vs. transversions in SNPs. Transitions are mutations within the same type of nucleotide: pyrimidine-pyrimidine mutations (C <-> T) and purine-purine mutations (A <-> G). Transversions are mutations from a pyrimidine to a purine or vice versa. Transitions occur at a higher rate because C, particularly when it is adjacent to a G i.e. CpG, can easily get methylated; once methylated, if deaminated it turns into U (equivalent to T), causing G->A on the other hand. (For instance, rs74315403, the SNP that causes fatal familial insomnia, is in fact a G -> A mutation on the protein coding strand, GAC (aspartate) -> AAC (asparagine), presumably originally caused by a methylation + deamination of the C on the non-coding strand). Note that there are twice as many possible transversions as transitions, which leads to a huge amount of confusion and disagreement as to whether Ti/Tv is supposed to be 0.5 or 1.0 in the absence of a biological bias towards transitions, i.e. whether it is a ratio of rates of mutations or a ratio of mutations, i.e. whether you need to multiply count(transitions)/count(transversions) by 2 (example of disagreement and discussion on biostars). After some research, I am going to side with the one web reference I’ve found who gives his source: Mark DePristo states that Ti/Tv should be 0.5 for false positives i.e. under randomness / absent biological forces, implying that it is not adjusted but rather simply the ratio of the number of transitions to the number of transversions. He cites Ebersberger 2002, who compares humans and chimpanzees and gives the raw count of Ti and Tv in Table 1, which works out to a Ti/Tv of 2.4 using raw counts. DePristo states that the Ti/Tv ratio should be about 2.1 for whole genome sequencing and 2.8 for whole exome, and that if it is lower, that means your data includes false positives caused by random sequencing errors, and you can calculate your error rate based on how much the false positives have dragged the overall mean down towards their mean of 0.5. (Aside: presumably the greater transition content of the exome is ultimately because the exome is under stronger selective pressure against missense mutations, whereas many transitions are tolerated as silent mutations because transitions in the third base of a codon rarely change the amino acid. There is also a possible connection with CpG islands that I don’t fully understand yet.)
- missingness simply means at what fraction of sample-site combinations you were unable to call a genotype. Often the inverse is stated instead: 2% missingness means a genotyping rate or call rate of 98%.
- ns/ss is, I *think*, the ratio of non-synonymous substitutions (per site) to synonymous substitutions (per site). If so, Wikipedia calls it the dN/dS ratio (see page on Ka/Ks ratio). I can’t find a good reference for what this value “should” be. Since synonymous substitutions are better tolerated by evolution, I’d expect the ratio to be below 1 (and indeed, Emond’s is 0.8), and that random errors would skew it upward.
- het/hom is the ratio of heterozygous to homozygous variant genotypes across all sample-site combinations. I can’t find a good web reference for what this “should” be, probably because it depends: populations with recent admixture will skew towards heterozygosity, populations with inbreeding will skew towards homozygosity.
- singletons are variants found in only one of your samples. Emond apparently had an average of ~300+ singletons per sample. I would expect this number to be higher if you have fewer samples (Emond had 91, I have 50) because, in the limit where you have one sample, every variant is a singleton. Knudsen 2009 uses the definition of singleton I’ve used here and points out that random errors in sequencing will very likely end up to be singletons, so an elevated number of singletons could indicate that errors are present. Frustratingly, I haven’t been able to find a good reference for what’s a “normal” number of singletons as a function of the number of samples, probably because it would depend on how genetically diverse your samples were, and how much of the genome you sequenced (whole genome vs. exome, and if exome, how many Mb your capture kit targeted). see 2012-10-22 update below re: “correct” number of singletons. (FYI: the term singleton has other uses in bioinformatics as well, such as a read whose mate didn’t align in paired-end sequencing — for instance see SeqAnswers).
- The final three figures are all different measures of depth. Depth, or coverage, is the number of reads overlapping a given base after alignment. There is code in the GATK exome pipeline to get average coverage of target and fraction of target covered at 8x or greater.
It’s probably good to compute all of these figures on your variants before proceeding to downstream analysis and modeling. If you notice that any numbers are far from what you’d expect, you probably have some errors upstream that are going to mess with your analysis. The PLINK/SEQ stats module can compute most of these measures for you, and a few others as well, in under a minute with a single command line:
pseq variants.vcf v-stats
If you want to separate out your cases and controls as Emond did, you’re probably best off just using a mask, as follows:
pseq variants.vcf v-stats --mask indiv=case1,case2,case3... pseq variants.vcf v-stats --mask indiv=control1,control2,control3...
PLINK/SEQ does not compute ns/ss or het/hom. Luckily these are pretty trivial. If you have your variants in a SQL database according to the table schemata from my GATK exome pipeline, here’s a query for het/hom:
select sum(case when variant_allele_count = 1 then 1 else 0 end)::numeric/sum(case when variant_allele_count = 2 then 1 else 0 end)::numeric as het_hom_ratio from sample_variants where alleles_genotyped = 2 ;
ns/ss is a slightly more of a pain — it’s not just the ratio of nonsynonymous variants to synonymous variants, it’s the ratio-of-the-ratios: nonsynonymous variants/sites to synonymous variants/sites. This query will take at least 3 minutes to run:
select ( sum(case when e.effect = 'NON_SYNONYMOUS_CODING' then variant_allele_count else 0 end)::numeric/
sum(case when e.effect = 'NON_SYNONYMOUS_CODING' then 1.0 else 0 end)::numeric ) /
( sum(case when e.effect = 'SYNONYMOUS_CODING' then variant_allele_count else 0 end)::numeric/
sum(case when e.effect = 'SYNONYMOUS_CODING' then 1.0 else 0 end)::numeric ) as ns_ss_ratio
from var06.sample_variants sv, (select vid, effect from var06.effects where effect in ('SYNONYMOUS_CODING','NON_SYNONYMOUS_CODING') group by vid, effect) e
where sv.vid = e.vid
;
You’ll notice the subquery allows multiple rows per variant, which I consider to be correct — if a variant can have either a synonymous or nonsynonymous effect depending on the transcript, it should count towards both categories.
One other thing you might be interested in is how many of your variant sites were sequenced at what depth (as opposed to how many of all sites). There are a few different ways to approach this:
-- number of variants where _all_ samples had depth of at least 8 select count(*) nvar from (select vid, min(dp) mindp, count(dp) ndp from var06.sample_variants sv group by vid) sub where sub.mindp >= 8 and ndp = 50 -- your number of samples ; -- number of variants where average depth was at least 8 select count(*) nvar from (select vid, avg(coalesce(dp::numeric,0.0)) avgdp from var06.sample_variants sv group by vid) sub where sub.avgdp >= 8 ; -- fraction of sample-site combinations with depth at least 8 select sum(case when dp >= 8 then 1.0 else 0.0 end)/count(*)::numeric fraction from var06.sample_variants sv ;
update 2012-10-17: as an alternative or supplement to PLINK/SEQ, vcftools stats modules can also compute most of these metrics, plus per-individual heterozygosity measurements (presumably to detect admixed or inbred individuals), Hardy-Weinberg Equilibrium by site, and a few others.
update 2012-10-22: Though I still can’t speak to how many singletons your samples “should” have, I did some calculations to see how it depends upon the number of samples. I have 50 samples, so I calculated the number of singletons in different-sized subsets of them to find a curve. Here’s how I did it:
First, I used a few lines of Python code to generate command line calls for PLINK/SEQ for 5 samples, 10 samples, etc.:
for j in range(5,50,5):
l = list(range(0,j))
print "pseq variants.final.vcf v-stats --mask indiv="+','.join(map(str,l))
This generates calls like this:
# 5 samples pseq variants.final.vcf v-stats --mask indiv=0,1,2,3,4 # 10 samples pseq variants.final.vcf v-stats --mask indiv=0,1,2,3,4,5,6,7,8,9
etc. I then ran PLINK/SEQ 50/5 = 10 times, and recorded the number of singletons each time. I then put my results into R:
samples = c(5,10,15,20,25,30,35,40,45,50) singletons = c(32563,43226,46598,48469,51513,53588,55512,56691,57393,58557)
And tried a few different models. The model with the most explanatory power (R2 of almost 99%) was singletons proportional to log number of samples:
> model <- lm(singletons ~ log(samples))
> summary(model)
Call:
lm(formula = singletons ~ log(samples))
Residuals:
Min 1Q Median 3Q Max
-1391.70 -496.50 52.08 295.42 1719.44
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 16419.8 1292.6 12.70 1.39e-06 ***
log(samples) 10895.0 404.4 26.94 3.88e-09 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 889.3 on 8 degrees of freedom
Multiple R-squared: 0.9891, Adjusted R-squared: 0.9877
F-statistic: 725.8 on 1 and 8 DF, p-value: 3.879e-09
I have 50 samples, and on the face of it, my number of singletons (58,557) had seemed rather elevated to me: almost 1200 singletons per person, compared to ~300 in Emond’s paper. Armed with this new model, I checked whether my results were really that different. Plugging in the slope and intercept from above, I can predict that if I had 91 samples (as Emond 2012 did), then I’d have (16419+10895*log(91))/91 = 720 singletons per sample. To adjust this a bit further, Emond states in online methods that three different exome capture kits were used, targeting 26, 32 and 34 Mb. She doesn’t state the relative proportions, so I just chose to assume that her singleton counts correspond to the middle value of 32 Mb. By contrast I am working with a little over 48 Mb, so I’d expect 1.5x as many singletons. 720/1.5 = 480. So the most appropriate comparison to Emond’s 300-odd singletons is a number around 480. While this is a bit elevated, it’s not nearly as bad as 1200. It might be partly that I have some spurious variant calls, and/or partly the result of a more diverse population.
update 2012-11-12: I discovered that PLINK/SEQ and snpEff compute the ts/tv ratio differently from each other. I suspect some of the difference is in whether variant, or sample-variant, is the unit of analysis. But neither product’s documentation explains exactly what it is doing. So I wrote my own SQL query for ts/tv to be sure that I know exactly what I’m getting:
create function nttype(nt varchar) returns char(1)
as $$
select case when $1 in ('C','T') then 'Y' when $1 in ('A','G') then 'R' else null end
$$ language sql
-- ts/tv ratio
select sum(case when nttype(v.ref) = nttype(v.alt) then 1.0 else 0.0 end)/sum(case when nttype(v.ref) <> nttype(v.alt) then 1.0 else 0.0 end) as tstv
from sample_variants sv, variants v
where v.vid = sv.vid
and alleles_genotyped = 2
-- and v.filter = 'PASS'
;
And once you’ve got this, it’s also easy to check for only coding regions if you want:
select sum(case when nttype(v.ref) = nttype(v.alt) then 1.0 else 0.0 end)/sum(case when nttype(v.ref) <> nttype(v.alt) then 1.0 else 0.0 end) as tstv
from sample_variants sv, variants v, (select vid, min(coding) coding from effects where coding <> '' group by vid) e -- if coding in any transcripts, consider it coding but count only once per vid
where v.vid = sv.vid
and v.vid = e.vid
and alleles_genotyped = 2
and e.coding = 'CODING'
and v.filter = 'PASS'
;