An introduction to Walker Jackson's fatal familial insomnia knock-in mice
 Read with caution! This post was written during early stages of trying to understand a complex scientific problem, and we didn't get everything right. The original author no longer endorses the content of this post. It is being left online for historical reasons, but read at your own risk. |
In 2009, Walker Jackson, then a postdoc in Susan Lindquist’s lab at MIT’s Whitehead Institute (now a group leader at DZNE) published his creation of fatal familial insomnia knock-in mice [Jackson 2009]. As Prion Alliance has recently decided to make testing candidate therapeutic molecules in these mice one of its research priorities, I’ve spent a lot of time educating myself on this mouse model and am going to document the key points in this post.
First, some background. Dr. Jackson’s primary purpose in creating these mice was to demonstrate experimentally what most prion scientists had already believed for decades: that prion infectivity is encoded in the conformation of misfolded PrP, and therefore can be generated spontaneously from genetic mutations. Lindquist’s lab had studied scrapie and CJD for years, so to demonstrate conclusively that prion infectivity could arise spontaneously from a genetic mutation, Jackson needed to be able to rule out the possibility that any mice in the experiment had simply acquired prion disease from prions left in the mouse cages from other experiments. That need motivated two decisions about the mouse model he generated: first, he chose FFI as the disease to study because FFI (in humans) was known to have phenotypes distinct from CJD and other prion diseases: insomnia, hyper- or hypothermia, astrogliosis and neuronal loss localized in the thalamus. Skeptics wouldn’t be able to argue that mice with these phenotypes had simply acquired prion disease from other mice or laboratory contaminants. Second, he engineered an additional change to the mice’s Prnp DNA sequence to introduce a species barrier between his mice and previous mice studied in the lab. He did so by introducing two humanizing mutations: L109M and V112M, thus completing the 3F4 epitope. That’s an antibody binding site on HuPrP believed to consist of the KTNMKHM at codons 106-112 inclusive [Lund 2007], of which the Ms at 109 and 112 are thought to be crucial. Jackson correctly suspected that introducing this epitope would make it more difficult for these mice to infect other mice or to become infected with rodent strains of scrapie.
The mouse line Jackson created is known as ki-3F4-FFI. ki stands for knock-in: these mice carry the 3F4 epitope and FFI mutation as engineered changes to what is otherwise mouse PrP. He also created control ki-3F4-WT mice as a control: mice with the 3F4 knock-in but no FFI mutation.
The frustrating thing about being a postdoc doing mouse experiments—or a rare disease advocate hoping for progress in drug testing—is that it takes years to get any results. To speed up results from the mouse model, Dr. Jackson bred the mice to be homozygous for the knock-in disease gene. To our knowledge, there’s never been a homozygous human with FFI, but there have been a handful of homozygous E200K CJD cases suggesting that homozygous mutations lead to an earlier age of onset [Simon 2000]. Jackson also experimented with different genetic backgrounds of mice. A mouse is not a mouse is not a mouse. Laboratory mice come in a variety of strains with names like “129/Ola” and “Black-6″. Those two strains both make appearances in today’s story, and are also pretty famous. Slate has published an awesome pop science article about the importance and pitfalls of Black-6 mice in research. 129/Ola mice have some history in prion research and among other things were used to create the first PrP knockout mice [Manson 1994].
These strains (like most of the mouse lines used in research) are highly inbred, which means exactly what it sounds like. Inbred lab mice are designed to be as genetically identical as possible so that experiments are more tightly controlled. Most inbred lines are at least 99% homozygous, with genomes often available for download—for most purposes you don’t need to sequence them yourself.
Breeding is necessary not only to perpetuate your mouse line but even just to get a knock-in (or transgenic) mutant in the first place. When you mutate embryonic stem cells to create a mouse model, you don’t get a purely mutant animal right away. You inject the mutated cells into a blastocyst and as it develops, parts of the mouse carry the mutation and parts don’t. This mouse is called a chimera. If the mutation winds up in the germline, you can breed the mouse and get offspring which carry the mutation in every cell in their bodies. Then you have to start crossing these mice to the genetic background that you want the mutation on, at a cost of about 9 or 10 weeks per generation at best. Below is a nice illustration of the general idea. In the illustration the cells are transgenic rather than knock-in but to my understanding the part of the process depicted in the diagram is the same either way.
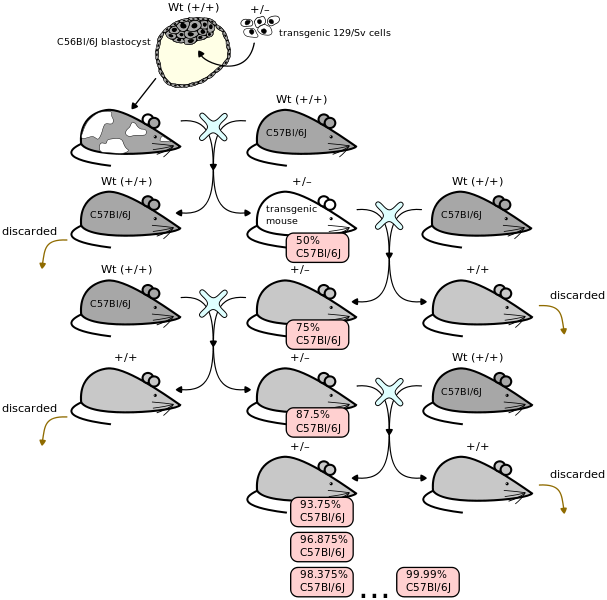
(Above image contributed to Wikimedia Commons by Seans Potato Business under GFDL)
For more background on this process, don’t miss this Flash animation by Jax. This especially helped me understand the nomenclature: when backcrossing to an inbred breed to get the new mutation onto a well-characterized background strain, the generations are termed N1, N2, … while when outcrossing to a different breed the generations are termed F1, F2, etc.
When Dr. Jackson first created the ki-3F4-FFI mice, he did so in embryonic stem cells from 129/Ola mice. The resulting chimeras were bred to C57Bl/6N-Tac aka Black-6 mice. The first generation of mice born with the mutation are referred to as F1. He then backcrossed the F1s to Black-6 mice again for an N1. Next he crossed the (heterozygous for the FFI gene) N1s with each other, resulting in N2s that were 25% 129/Ola and 75% Black-6 and homozygous for the knock-in gene. These N2s were then bred to each other to produce the mice analyzed in the 2009 Neuron paper.
As time went on, Dr. Jackson also tried further backcrossing these ki-3F4-FFI mice with the Black-6 background, but discovered that after a few more generations the phenotype became very difficult to discern. The mice got sick late and mildly or not at all, and it was difficult to detect reactive gliosis in the histological sections.
So, starting from an earlier generation with less Black-6 in it, he started breeding the mice over to the 129/SvJae strain (see Jax’s 129 mice; apparently this strain is actually pretty different from 129/Ola). He’s done a good deal of analysis on the N2 mice from this line (so ~75% 129/SvJae) and they appear to get FFI symptoms earlier, more robustly and more reliably. Reactive gliosis is so visible that their brain sections can practically be genotyped at a glance.
Though less study has been done on the later generations which are as much as 96% 129/SvJae, it appears that they also have an earlier and more reliable phenotype. At Prion Alliance we are working on reaching out to other scientists interested in testing candidate therapeutics in these mice—particularly therapeutics that have already shown a significant effect in scrapie mice.
While the mice have a phenotype recognizable as FFI and distinct from other prion diseases, more work needs to be done to determine exactly how best to measure and characterize this phenotype. In the 2009 study, Jackson used three different phenotyping approaches. First, he set up cameras in the home cages of 22 FFI mice and 14 control mice and used Automated Mouse Behavioral Analysis (AMBA) to measure the time the mouse spent in various behaviors: sleeping, rearing, hanging from a bar, twitching, etc. Second, he monitored the core temperature of five FFI and five control mice using iButtons surgically embedded inside the mouse (core temperature was necessary because surface temperature fluctuates naturally and doesn’t indicate disease). Third, after the mice had died, he examined hematoxylin and eosin-stained sagittal sections to identify reactive gliosis, neuronal loss and vacuoles. These approaches were perfectly serviceable for his purposes at the time, showing that most mice exhibited hyper- or hypothermia, slept less and moved restlessly more often, and had neurodegeneration concentrated in the thalamus with some mice showing significant effects in the cerebellum and some elsewhere as well. All of these features conclusively identified the phenotype as belonging to FFI and not CJD or scrapie.
Despite all these phenotypic features, it is not clear that the mice actually die of FFI, and their lifespans are not especially shorter than wild-type mice. So age at death will not be a viable readout for a therapeutic study, and not just for this reason. Other reasons include: not wanting to wait ~two years to get results, and wanting to limit positive results to molecules that delay disease rather than extending disease.
When I spoke with Dr. Jackson recently, he noted the phenotypes he measured for the 2009 paper would not necessarily be suitable as readouts for a study of candidate therapeutics. The AMBA data were interesting but showed a number of false positive significant differences between the control (ki-3F4-WT) and true wild-type mice (see Supplemental Figure 5). They might still work as a readout but Dr. Jackson doesn’t feel he has done enough analysis on this to be able to say for certain. In particular, he hasn’t done AMBA analysis on the more recent generations of primarily 129/SvJae mice. The iButtons stored temperature data locally and it could not be accessed until after the mice had died. Newer telematic models are available to provide real-time core temperature data. Real-time core temperature data could be one valuable source of phenotypic information for FFI mice in a therapeutic study, but given that the previous study only looked at temperatures for five mice, the mean and variance of the age of onset for temperature dysregulation are not well-established yet.
By far the most robust phenotype observable in the ki-3F4-FFI mice is reactive gliosis. Reactive gliosis in FFI is the hypertrophy (i.e. enlargement) and/or proliferation of astrocytes, which are glial cells (hence “gliosis”) in response to disease stress (hence “reactive”). Reactive gliosis is readily identifiable in histological sections of FFI mouse brains, particularly in the thalamus. But again, therapeutic studies will require a real-time readout while the mice are alive. Dr. Jackson suggested that the best readout for reactive gliosis would be live animal imaging of luciferase under the GFAP promoter.
What does that mean? GFAP, or glial fibrillary acidic protein, is a protein which is uniquely expressed in glial cells and not in neurons. During prion disease, not only do astrocytes multiply in number, GFAP is also transcriptionally upregulated. For both of these reasons, the amount of GFAP in a mouse brain is a very reliable indicator of prion disease phenotype. Traditionally, post-mortem histological samples are stained with a fluorescent antibody for GFAP. But live animal imaging is now possible as well. Others have already engineered mice expressing firefly luciferase (a protein which catalyzes a bioluminescent reaction and is widely used in assays) under the GFAP promoter (you can buy these mice from JAX). So when GFAP expression increases, more luciferase is produced. Some of the wavelengths of light emitted by the reaction that luciferase catalyzes are infrared and so pass through the mouse’s tissues and can be viewed in the live mouse using infrared cameras. (For a review of several published studies using this technology, see Dothager 2009) So basically, put on your infrared goggles, and you’ll see glowing FFI mice and not-so-glowing control mice. The luciferase-under-GFAP mice are already available (for sale online from JAX) and would just need to be bred to the ki-3F4-FFI mice. Of course, as Mendel knows, it will take two generations to get mice homozygous for both luciferase-under-GFAP and ki-3F4-FFI. This cross-breeding could be hired out to companies if desired.
Live animal imaging of reactive gliosis promises to probably be the best phenotypic readout for a therapeutic study on FFI mice. But since it hasn’t been done yet, we don’t yet have solid data on the mean and variance in age of onset of this phenotype. These are important numbers for anyone designing a mouse study: you need to know going in that you’ve got enough mice to get a statistically significant result for the effect size you expect. I asked Dr. Jackson for his subjective assessment of the age of onset and its variability. He said that in the current 129/SvJae background, about half the mice seem to show reactive gliosis by 12 months, with most occurring within a range of a couple of months around that. He said 60 weeks would probably be enough time to allot for a study to run to conclusion. He pointed out to me, which I hadn’t realized before, that intracerebral injections of scrapie (which are the leading brand of prion mouse model, used in nearly all of the studies I’ve reviewed on this blog) represent not only the best (fastest, least variable, most phenotypically reliable) mouse model of prion disease, but possibly the best mouse model of any neurological disease (possibly second to stroke and just ahead of R6/2 for Huntington’s). The studies of therapeutics in scrapie mice that I’ve reviewed on this blog have used 6, 10 or 15 mice per treatment group (see Scutellaria lateriflora, statins and styryls/tricyclics respectively); whereas for the FFI mice, given the greater variance in age of onset, 20 mice per treatment group will probably be necessary. 20 (or more) mice per group would be consistent with the numbers from the only other therapeutic study in a genetic model that we are aware of— Mastrianni’s work with rapamycin in transgenic GSS mice.
Dr. Jackson was also kind enough to share with me some unpublished data on reactive gliosis in the mice so that I could draw my own conclusions about phenotypic variability. I’ll be taking a closer look at that data in a shortly forthcoming power analysis post.