Is PRNP mRNA alternatively spliced?
In my last post I discussed the possible mechanisms by which a small molecule could reduce the expression level of a single gene of interest, with minimal effect on other genes. It appears that there exists decent proof of principle for targeting splicing with a small molecule, thanks in large part to Novartis’ recent success developing LMI-070 [Palacino 2015], a splice-modifying drug being tested in a clinical trial for spinal muscular atrophy. Therefore I set out to learn whether there is anything unique about the splicing of PRNP mRNA that we might hope to target therapeutically.
As a quick review of splicing vocabulary, the end of one exon and the beginning of an intron is referred to as the donor or 5’ splice site (named for being at the 5’ end of the intron), the end of an intron and beginning of the next exon is the acceptor or 3’ splite site (again, at the 3’ end of the intron). A few bases at splice sites are highly stereotypical:
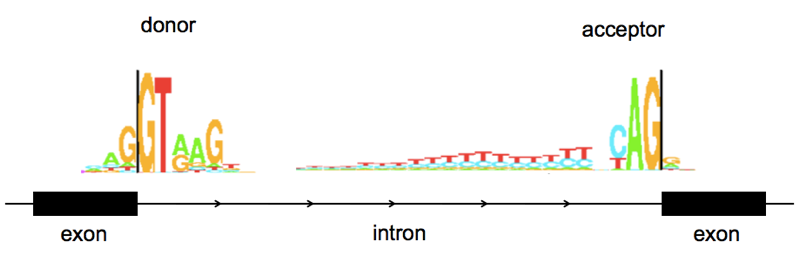
Above: sequence logos for human splice donor and acceptor sites, from Tom Schneider’s lab at NIH here. Note that these are from a rather old paper [Stephens & Schneider 1992] and are based on just ~1700 splice sites. I have used them here because as works of a U.S. federal government employee, they are in the public domain. I couldn’t find a newer sequence logo that was Open Access, but if you look at more recent papers (e.g. [Roca 2013]) nothing seems to have changed.
Human PRNP only has two exons, and therefore just one splice donor and one acceptor, so there isn’t as much to work with here as you might have with some genes. It’s on chromosome 20 and occupies a total of ~15kb of genomic sequence, most of which is the intron:
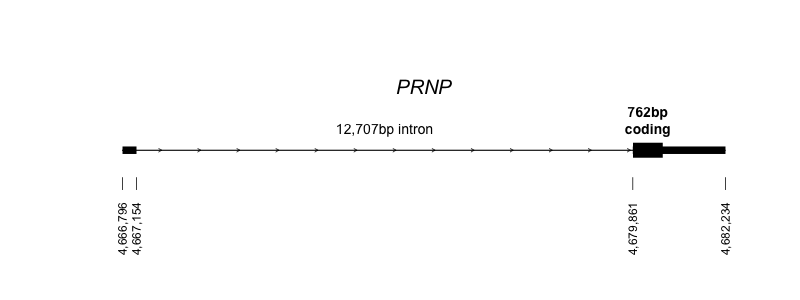
Above: the structure of the PRNP gene on chromosome 20, in hg19/GRCh37 coordinates (code). Though you can’t see it in the diagram above because it’s so short, there is a tiny bit of sequence in exon 2 before the coding sequence begins.
As discussed in my CRISPR post, the splice acceptor of PRNP exon 2 is something you might not want to tinker with. In the Nagasaki line of PrP knockout mice [Sakaguchi 1995], they deleted the orthologous splice acceptor in addition to the gene’s protein-coding sequence, and this resulted in expression of a fusion transcript under the PrP promoter and 5’UTR but with the coding sequence of Dpl (PRND in humans), the next gene downstream. The resulting ectopic expression of Dpl protein was neurotoxic [Sakaguchi 1996, Nishida 1999, Moore 1999, Li 2000, Silverman 2000, Rossi 2001, Moore 2001].
That said, we don’t know if the same thing would happen in humans. And interfering with splicing with a small molecule might be different than deleting the splice acceptor in genomic DNA. And finally, the neurotoxicity of Dpl expression can be rescued by PrPC expression, and you’re unlikely to be lucky enough to find a drug that knocks down PrP by 100%, so in all likelihood there will always still be some PrP around. For all of these reasons, I think it is still worth discussing the possibility of looking for compounds targeting PRNP splicing.
The original paper characterizing a human PRNP cDNA [Liao 1986] said nothing about splicing. The cDNA used in a later, more detailed, characterization of the gene [Puckett 1991] was the product of a highly canonical splicing event: CAG|GTAAA at the donor, and TTTTGCAG|AG at the acceptor. This is similar to the sequence motif found in nearly all human splice sites (see sequence logos above).
Yet if you look in UCSC Genome Browser at the regions of the donor and acceptor you will find there are several RefSeq transcripts for PRNP displayed, which differ by several bases in the exact cut site for the exon/intron and intron/exon boundaries:
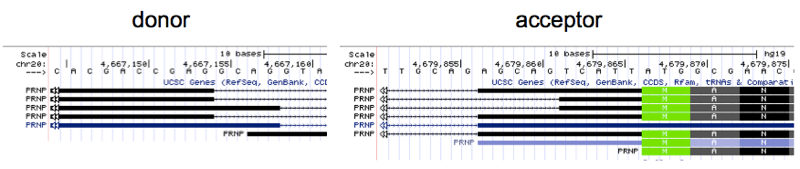
I stared at these for a long time trying to decipher whether they actually represent distinct cDNA sequences, or are merely alignment artifacts. They are in fact different. For instance, the mRNA sequence implied by the second-from-top transcript displayed above contains the string CGAGTCATTA, while the third-from-top contains GCAGTCATTA instead; this difference can only be explained by a different splice donor location. Does this reflect variability in splicing within one human genome, or between individuals? According to the 1000 Genomes browser none of the 2504 humans in 1000 Genomes seem to have any variants at the splice donor.
My understanding is that transcripts deposited in RefSeq come mostly from older technologies, so I also wanted to check if the differences in splicing can be observed in slightly more modern RNA-seq data. Open access RNA-seq BAMs are shockingly difficult to find on the internet, but I did turn to the Human BodyMap dataset, for which raw BAMs are available via FTP. I downloaded the brain BAM, and to identify reads that confidently span the exon-exon junction of PRNP mRNA, I samtools viewed the region of interest and greped for sequences unique to exon 1 and exon 2:
samtools view GRCh37.HumanBodyMap.brain.1.bam 20:4667151-4679881 | awk -v FS="\t" '{print $10}' | grep TCTCCTCACGACCG | grep ATGGCGAAC
I obtained 46 reads, which I copied into a text file and aligned manually based on the unique sequences upstream and downstream of the putative alternative splicing region. Here’s what I saw:
NNTGACAGCCGCGGCGCCGCGAGCTTCTCCTCTCCTCACGACCGAG AGCAGTCATTATGGCGAACCTTGGCTGCT
CTGACAGCCGCGGCGCCGCGAGCTTCTCCTCTCCTCACGACCGAG GCAGAGCAGTCATTATGGCGAACCTTCGCT
CAGCCGCGGCGCCGCGAGCTTCTCCTCTCCTCACGACCGAG AGCAGTCATTATGGCGAACCTTGGCTGCTGGATG
CNGCCGCGGCGCCGCGAGCTTCTCCTCTCCTCACGACCGAG GCAGAGCAGTCATTATGGCGAACCTTGGTTGCTG
NNGCGGCGCCGCGAGCTTCTCCTCTCCTCACGACCGAG AGCAGTCATTATGGCGAACCTTGGCTGCTGGATGCTG
NNGCGGCGCCGCGAGCTTCTCCTCTCCTCACGACCGAG AGCAGTCATTATGGCGAACCTTGGCTGCTGGATGCTG
CCGCGGCGCCGCGAGCTTCTCCTCTCCTCACGACCGAG AGCAGTCATTATGGCGAACCTTGGCTGCTGGATGCTG
NNCGGCGCCGCGAGCTTCTCCTCTCCTCACGACCGAG AGCAGTCATTATGGCGAACCTTGGCTGCTGGATGCTGG
CGCGGCGCCGCGAGCTTCTCCTCTCCTCACGACCGAG GCAGAGCAGTCATTATGGCGAACCTTGGCTGCTGGATG
CGCGGCGCCGCGAGCTTCTCCTCTCCTCACGACCGAG GCAGAGCAGTCATTATGGCGAACCTTGGCTGCTGGATG
CGCGGCGCCGCGAGCTTCTCCTCTCCTCACGACCGAG AGCAGTCATTATGGCGAACCTTGGCTGCTGGCTGCTGG
CGCGGCGCCGCGAGCTTCTCCTCTCCTCACGACCGAG GCAGAGCAGTCATTATGGCGAACCTTGGCTGCTGGATG
CGCGGCGCCGCGAGCTTCTCCTCTCCTCACGACCGAG AGCAGTCATTATGGCGAACCTTGGCTGCTGGATGCTGG
CGCGGCGCCGCGAGCTTCTCCTCTCCTCACGACCGAG AGCAGTCATTATGGCGAACCTTGGCTGCTGGATGCTGG
CGCGGCGCCGCGAGCTTCTCCTCTCCTCACGACCGAG GCAGAGCAGTCATTATGGCGAACCTTGGCTGCTGGATG
CGCGGCGCCGCGAGCTTCTCCTCTCCTCACGACCGAG GCAGAGCAGTCATTATGGCGAACCTTGGCTGCTGGATG
CGCGGCGCCGCGAGCTTCTCCTCTCCTCACGACCGAG AGCAGTCATTATGGCGAACCTTGGCTGCTGGATGCTGG
CGCGGCGCCGCGAGCTTCTCCTCTCCTCACGACCGAG GCAGAGCAGTCATTATGGCGAACCTTGGCTGCTGGATG
CGCGGCGCCGCGAGCTTCTCCTCTCCTCACGACCGAG AGCAGTCATTATGGCGAACCTTGGCTGCTGGATGCTGG
CGGCGCCGCGAGCTTCTCCTCTCCTCACGACCGAG GCAGAGCAGTCATTATGGCGAACCTTGGCTGCTGGATGCT
CGGCGCCGCGAGCTTCTCCTCTCCTCACGACCGAG GCAGAGCAGTCATTATGGCGAACCTTGGCTGCTGGATGCT
NACGCCGCGAGCTTCTCCTCTCCTCACGACCGAG TCATTATGGCGAACCT
GGCGCCGCGAGCTTCTCCTCTCCTCACGACCGAG TCATTATGGCGAACCT
GGCGCCGCGAGCTTCTCCTCTCCTCACGACCGAG AGCAGTCATTATGGCGAACCTTGGCTGCTGGATGCTGGTTC
GGCGCCGCGAGCTTCTCCTCTCCTCACGACCGAG GCAGAGCAGTCATTATGGCGAACCTTGGCTGCTGGATGCTG
NNCGCCGCGAGCTTCTCCTCTCCTCACGACCGAG GCAGAGCAGTCATTATGGCGAACCTTGGCTGCTGGATGCTG
GNCGCCGCGAGCTTCTCCTCTCCTCACGACCGAG AGCAGTCATTATGGCGAACCTTGGCTGCTGGATGCTGGTTT
GNCGCCGCGAGCTTCTCCTCTCCTCACGACCGAG AGCAGTCATTATGGCGAACCTTGGCTGCTGGATGCTGGTTC
GTCGCCGCGAGCTTCTCCTCTCCTCACGACCGAG AGCAGTCATTATGGCGAACCTTGGCTGCTGGATGCTGGTTC
GCGCCGCGAGCTTCTCCTCTCCTCACGACCGAG GCAGAGCAGTCATTATGGCGAACCTTGGCTGCTGGATGCTGG
GCGCCGCGAGCTTCTCCTCTCCTCACGACCGAG GCAGAGCAGTCATTATGGCGAACCTTGGCTGCTGGATGCTGG
GCGCCGCGAGCTTCTCCTCTCCTCACGACCGAG GCAGAGCAGTCATTATGGCGAACCTTGGCTGCTGGATGCTGG
NNCCGCGAGCTTCTCCTCTCCTCACGACCGAG GCAGAGCAGTCATTATGGCGAACCTTGGCTGCTGGATGCTGGT
GCCGCGAGCTTCTCCTCTCCTCACGACCGAG AGCAGTCATTATGGCGAAC
GCCGCGAGCTTCTCCTCTCCTCACGACCGAG AGCAGTCATTATGGCGAAC
CCGCGAGCTTCTCCTCTCCTCACGACCGAG GCAGAGCAGTCATTATGGCGAACCTTGGCTGCTGGATGCTGGTTC
CGGCGAGCTTCTCCTCTCCTCACGACCGAG AGCAGTCATTATGGCGAACCTTGGCTGCTGGATGCTGGTTCTCTT
CCGCGAGCTTCTCCTCTCCTCACGACCGAG AGCAGTCATTATGGCGAACC
CGCGAGCTTCTCCTCTCCTCACGACCGAG AGCAGTCATTATGGCGAACCT
CGCGAGCTTCTCCTCTCCTCACGACCGAG GCAGAGCAGTCATTATGGCGAACCTTGGCTGCTGGATGCTGGTTCT
CGCGAGCTTCTCCTCTCCTCACGACCGAG AGCAGTCATTATGGCGAACCT
NNGAGCTTCTCCTCTCCTCACGACCGAG GCAGAGCAGTCATTATGGCGAACCTTGGCTGCTGGATGCTGGTTCTC
CGAGCTTCTCCTCTCCTCACGACCGAG GCAGAGCAGTCATTATGGCGAAC
CGAGCTTCTCCTCTCCTCACGACCGAG GCAGAGCAGTCATNATGGCGAAC
AGCTTCTCCTCTCCTCACGACCGAG GCAGAGCAGTCATTATGGCGAACCT
AGCTTCTCCTCTCCTCACGACCGAG GCAGAGCAGTCATTATGGCGAACCT
After staring at this alignment, it became clear that there are in fact two distinct splicing events going on here. Of the 46 reads displayed here, 21 (47%) contain a CGAGGCAG sequence consistent with splicing at the canonical GT donor (herafter “type 1”). 23 (50%) contain a CGAGAGCAG sequence (“type 2”), and 2 (4%) contain a CGAGTCATTA sequence (“type 3”), both of which can only be explained by non-canonical splicing.
To figure out what was going on here, I lined up the sequences flanking the general region of the splice donor and acceptor, and delineated all possible splicing events that could have led to the observed reads being generated:
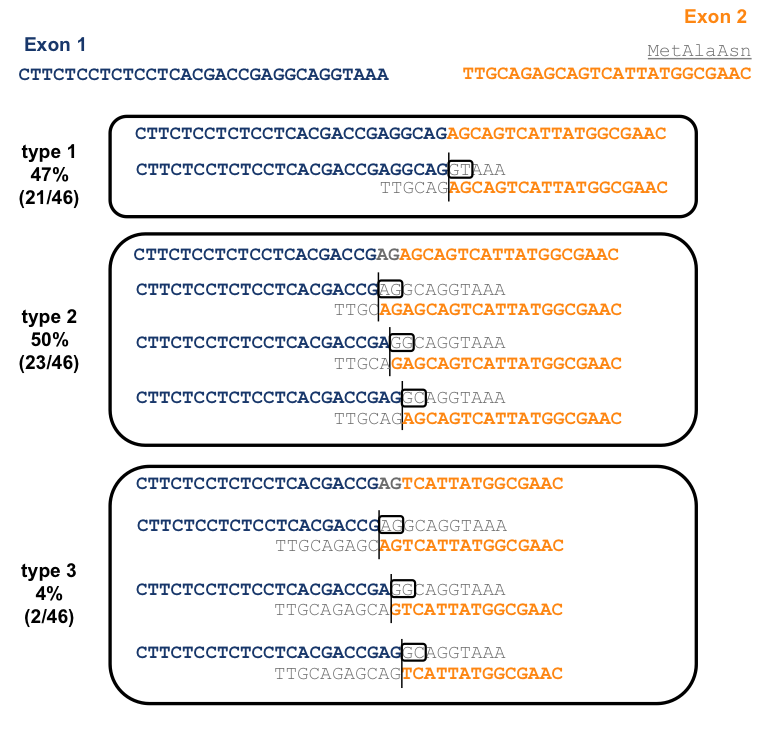
Because an AG sequence is repeated near the beginning and end of intron 1, there is ambiguity in the alignment for all three of these types reads. However, for type 1, those reads that contain a CGAGGCAG, one alignment is by far the parsimonious one, so it’s the only one I have depicted here. This alignment corresponds to a CAG|GTAAA donor and TTTTGCAG|AG acceptor just as described in [Puckett 1991] and is absolutely canonical (see above). For types 2 and 3 reads, containing CGAGAGCAG and CGAGTCATTA respectively, there are three possible alignments. The +1 and +2 positions of the donor must be either AG, GG, or GC in each case, all of which are non-canonical. Of the three options, I would favor the GC alignment (the third option shown for each read type above) because at least in this case, the splice acceptor of CAG|AG is fairly canonical, whereas for the other two options both the donor and acceptor are highly non-canonical.
It would appear, then, that just over half of RNA-seq reads for PRNP in this sample are coming from this unusual splice donor sequence of CGAG|GCAGG. I asked a colleague who studies splicing, and learned that GC splice donors are a rare but recognized entity in the human genome, accounting for ~0.56% of donors [Burset 2000]. This makes GC, albeit by a longshot, the second-most common splice donor motif.
Does this GC splicing event in PRNP occur in all humans, or does it just happen to occur in Human BodyMap? I didn’t find any other good public RNA-seq datasets, but a collaborator allowed me to have a look at a few BAMs from some human cortex RNA-seq samples. Here’s what I found.
samtools view $bampath 20:4667151-4679881 | awk -v FS="\t" '{print $10}' | grep TCTCCTCACGACCG | grep ATGGCGAAC | grep CGAGGCAG | wc -l # type 1
samtools view $bampath 20:4667151-4679881 | awk -v FS="\t" '{print $10}' | grep TCTCCTCACGACCG | grep ATGGCGAAC | grep CGAGAGCAG | wc -l # type 2
samtools view $bampath 20:4667151-4679881 | awk -v FS="\t" '{print $10}' | grep TCTCCTCACGACCG | grep ATGGCGAAC | grep CGAGTCATTA | wc -l # type 3
| BAM | type 1 | type 2 | type 3 | noncanonical |
|---|---|---|---|---|
| #1 | 31 | 32 | 0 | 51% |
| #2 | 144 | 92 | 2 | 39% |
| #3 | 206 | 159 | 1 | 44% |
| #4 | 136 | 111 | 1 | 45% |
| #5 | 161 | 68 | 2 | 30% |
So on average, somewhere just under half of the reads spanning the PRNP splice junction come from noncanonically spliced transcripts with a GC donor.
Does this mean that PRNP splicing could be targeted therapeutically to reduce expression? My immediate reaction is it doesn’t look too promising. If only about half of copies of PRNP mRNA are spliced this way, then even total inhibition of GC splicing would only reduce expression by about half. Meanwhile, if this motif is found in 0.56% of donors, such inhibition would probaly have undesired effects on hundreds of transcripts. On the other hand, the GA|GT donor motif targeted by LMI-070 is found in 2.6% of exons, and yet that therapeutic up-regulation of that splicing event was apparently well-enough tolerated in animals to lead to a clinical trial in humans. So I don’t totally rule out the possibility of targeting splicing, but until I learn more, I probably won’t think of it as a high priority.
