Population Substructure, Part II
This post is a sequel to How (and why) to create population covariates using 1000 Genomes data. Here are four more things you can and probably should do in the population substructure analysis started in that post.
1. Update on MDS plotting: use only common variants to avoid bias from which rare variants are included
In the previous post I described a method for combining your data with 1000 Genomes and then computing IBD on the whole set in order to generate an MDS plot. One problem I’ve discovered with this approach is as follows. The method I described will calculate IBD based on all variants that were called in 1000 Genomes and in your samples. My samples are much less numerous– I have 50 of my own samples and 629 1000 Genomes samples– and this introduces bias regarding rare variants. In particular, any variant will be included if it shows up as at least a singleton in both datasets. This means it must have allele frequency of 2% in my data and 0.15% in 1000 Genomes. This filter enriches the resulting list of SNPs for my own samples’ rare variation, comparatively depleting it for 1000 Genomes rare variation. (A hypothetical variant with 1% frequency in 1000 Genomes but not seen in my data would not be included).
When I originally plotted my data with 1000 Genomes, my samples clustered with each other but did not overlap with the 1000 Genomes clusters. At first I thought this might just indicate my samples came from populations not represented in 1000 Genomes (which seems plausible– 1000 Genomes still has no Southern or Central Africans, Pacific Islanders, Arabs, Central Asians, the list goes on) but when I plotted SNP array data for some of the same individuals with 1000 Genomes (see step 2 below) I found that they clustered tightly with 1000 Genomes populations. Hmmm. After thinking about it more, I realized that the enrichment of my samples’ rare variation makes my samples look more different from 1000 Genomes than they actually are. This causes my samples to cluster just a little ways away from the 1000 Genomes population that they really ought to belong to. So I applied a MAF filter:
plink --file variants.subset --freq --out variants # calculate allele frequency based on my variants plink --file all --read-freq variants.frq --maf .10 --all --genome --out all_maf10 # calculate genome file for MAF > 10% in my samples plink --file all --read-genome all_maf10.genome --cluster --mds-plot 2 --out all_mds_2_maf10 # calculate MDS plot based on only the MAF > 10% variants
And immediately, my samples clustered tightly with various 1000 Genomes populations and matched almost exactly the pattern seen in the SNP array data. Problem solved.
2. Validation with data from other studies
Some of my samples had been previously genotyped in a SNP array study, which provided the chance to validate that the variants I called with the GATK pipeline on exome data. Because the two studies used different sample IDs, I needed a match table. We had a match table lying around in some master Excel file of all samples we’ve genotyped, but just in case that file were to have any errors in it, I deemed it safest to create my own match table based on the actual genetic similarity between samples.
First, I extracted the relevant SNPs:
grep -o 'rs[0-9]*' original.snparray.data.map | sort | uniq > snparray.snplist.dedup grep -o 'rs[0-9]*' exome.data.map | sort | uniq > exome.snplist.dedup cat exome.snplist.dedup snparray.snplist.dedup | sort | uniq -d > shared.snplist
And after trying once to combine the files I got a PLINK error as usual and a shared.missnp output file, so then I excluded those SNPs on the second try:
plink --file original.snparray.data --extract shared.snplist --exclude shared.missnp --recode --out snparray.subset plink --tfile exome.data --extract shared.snplist --exclude shared.missnp --recode --out exome.subset plink --file snparray.subset --merge exome.subset.ped exome.subset.map --recode --out snparray-exome plink --file snparray-exome --all --genome --out snparray-exome sort snparray-exome.genome -k10 -r > snparray-exome.genome.sorted # sort on k10, the pi_hat field
This file then shows the pairs with highest pi_hat, a measure of relatedness. I opened it up and sure enough, the top hits were SNP array samples matching to the same individual’s exome data, with pi_hat of .98 or better. I created a match table based on this information. At first I tried to just write out a .TFAM file directly from PostgreSQL, but for yet-undiagnosed reasons that didn’t work with PLINK, so finally I wrote out the match table named recoded.txt and followed the PLINK documentation for updating individual information with the --update-ids command:
plink --file original.snparray.data --update-ids recoded.txt --extract shared.snplist --exclude shared.missnp --recode --out snparray.subset.with.exome.ids
And then compared the two datasets in merge mode 7, so that PLINK would report the discrepancies between the two datasets only where the sample was genotyped in both datasets:
plink --file snparray.subset.with.exome.ids --merge exome.subset.ped exome.subset.map --merge-mode 7 --out snparray.exome
This writes out a file, snparray.exome.diff, listing all the sample-site combinations which were genotyped in both datasets and where the genotypes do not agree. The log file also tells you the overall concordance rate:
Of 174260 overlapping SNPs, 145580 were both genotyped and 145259 were concordant Concordance rate is 0.997795
This exome data matched the SNP array data about 99.8%. What’s nice is, I was able to do this with two different exome sequencing outputs (at this point I’ve run the GATK pipeline several times, with different combinations of settings, reference genomes, pre-alignment trimming of reads or not) and see which accorded most closely with the SNP array data. Because this particular SNP array was genome-wide, there is only a small overlap (about 8500 SNPs) with the exonic variants I called in my exome sequencing, but still, arriving at the same answer through two completely different genotyping technologies definitely helps to build confidence in the data.
3. Detecting familial relationships – continued
In the earlier post I mentioned the need to calculate IBD on a homogenous sample in order to reliably detect familial relationships among your samples. All of my samples clustered pretty clearly with either the European, Asian or African cluster from 1000 Genomes, so I coarsely sectioned them into those three groups in order to re-calculate the .genome files within these groups:
copy (
select m.fid, m.iid
from all_mds_2_maf10 m left outer join kg_metadata k
on m.iid = k.coriell_sample_id
where k.population in ('GBR','CEU','FIN','TSI')
or (m.iid ~ '[0-9]*' and c1::numeric < 0 and c2::numeric > 0)
) to '/your/path/here/euro_list.txt';
copy (
select m.fid, m.iid
from all_mds_2_maf10 m left outer join kg_metadata k
on m.iid = k.coriell_sample_id
where k.population in ('CHS','CHB','JPT')
or (m.iid ~ '[0-9]*' and c1::numeric < 0 and c2::numeric < 0)
) to 'your/path/here/asian_list.txt';
copy (
select m.fid, m.iid
from all_mds_2_maf10 m left outer join kg_metadata k
on m.iid = k.coriell_sample_id
where k.population in ('ASW','LWK','YRI')
or (m.iid ~ '[0-9]*' and c1::numeric > 0)
) to 'your/path/here/african_list.txt';
And, conscious of the bias introduced by rare variation as described in point 1 above, I filtered to only use variants of MAF 10% or higher in the genome calculations:
plink --file all --maf .10 --genome --keep african_list.txt --out africans plink --file all --maf .10 --genome --keep asian_list.txt --out asians plink --file all --maf .10 --genome --keep euro_list.txt --out europeans
Then I read the resulting .genome files into PostgreSQL and compiled them into one table:
drop table if exists genome;
create table genome as select *, 'africans'::varchar as population from africans_genome;
insert into genome select *, 'asians'::varchar as population from asians_genome;
insert into genome select *, 'europeans'::varchar as population from europeans_genome;
And then looked to see if my genome calculations had validated the known relationships noted in the 1000G metadata:
select g.iid1, g.iid2, g.z0, g.z1, g.z2, g.pi_hat, k1.population, k2.population, k1.family, k1.type__unrel_duo_trio_, k1.relationship_, k2.family, k2.type__unrel_duo_trio_, k2.relationship_
from genome g left outer join kg_metadata k1 on g.iid1 = k1.coriell_sample_id left outer join kg_metadata k2 on g.iid2 = k2.coriell_sample_id
where pi_hat > .125 -- arbitrary cutoff to find high pi_hat
order by pi_hat desc;
And indeed, two known relationships validated quite well (I’ve bolded the z0, z1 and z2 columns):
"HG00153";"HG00158";0.0989;0.8867;0.0144;0.4578;"GBR";"GBR";"";"duo";"Child";"";"duo";"Mother" "HG00146";"HG00147";0.3051;0.4758;0.2191;0.4570;"GBR";"GBR";"";"duo";"Sibling";"";"duo";"Sibling"
And I also discovered strong evidence of other relationships not annotated in the 1000G metadata:
"HG00578";"HG00635";0.2471;0.5186;0.2344;0.4937;"CHS";"CHS";"SH052";"trio";"mother";"SH071";"trio";"mother" "NA20526";"NA20792";0.3217;0.4235;0.2547;0.4665;"TSI";"TSI";"";"unrel";"";"";"unrel";"" "NA19381";"NA19382";0.1211;0.8457;0.0332;0.4561;"LWK";"LWK";"";"unrel";"";"";"unrel";"" "NA19445";"NA19453";0.1183;0.8817;0.0000;0.4409;"LWK";"LWK";"";"unrel";"";"";"unrel";"" "NA19434";"NA19444";0.3957;0.4234;0.1809;0.3926;"LWK";"LWK";"";"unrel";"";"";"unrel";""
HG00578 and HG00635, two mothers from different CHS (Southern Chinese) trios, appear to be sisters. There’s almost a perfect 25-50-25 split on IBD 0/1/2. Two Tuscans (NA20526 and NA20792) and two Luhya (NA19434 and NA19444) also looked pretty strongly to be siblings, and two other pairs of Luhya people (NA19381 & NA19382 and NA19445 & NA19453) looked to be parent-child.
A Google search confirmed that I’m not the only one who has noticed these pairs. Jim Nemesh did a study of cryptic relatedness in the 1000G data summarized in documents now hosted at EBI. His cryptic relatedness summary reports the Tuscan pair, NA20526 and NA20792 as siblings, and his cryptic relations clusters report reports the other four relationships I listed above. The analysis is described further in the readme and powerpoint.
That’s about all I was able to validate, because no other duos or trios are complete in the August 2010 1000 Genomes release. It took a while to figure this out because the metadata is not totally consistent yet– you’ll note that the mother/child and sibling/sibling duos validated above do not have the family field filled in, and the relationship_ field is sometimes capitalized and sometimes not. But if you check:
select *
from kg_metadata
where relationship_ in ('child','Child')
and coriell_sample_id in (select iid1 from genome);
You’ll find only three ‘child’ individuals, and on further investigation, two do not have parents in this data release. And if you look for where relationship_ in ('sibling','Sibling'), the sibling due validated above are the only one. So indeed, my analysis successfully validated all of the known immediate family relationships that were present in the August 2010 1000 Genomes release. This gives me confidence that my genome calculations will be accurate with respect to any relationships among my own samples.
Important note: because the family field is not yet consistently used and most trios are not complete, if you throw in a where k1.family = k2.family condition, you’ll find just a list of people’s grandparents and parents i.e. people who are not immediately blood-related to each other, which, not surprisingly when you think about it, often appear distantly related to one another, with z1+z2 of ~4%. Leave this condition out in order to find the parent/child and sibling/sibling relations mentioned above.
Troublingly, I found a lot of cryptic relatedness– tens of pairs of 1000 Genomes people who aren’t labeled as related to each other have about 40% IBD-1, and a couple of individuals in particular had 60% IBD-1 with many people. Some of this may be an indicator that my European/Asian/African groupings were a bit on the coarse side, causing people from the same subpopulation to look excessively related to each other. But Jim Nemesh’s powerpoint on cryptic relatedness in 1000G finds plenty of such combinations as well, supporting the idea that this is really just a feature of the data. The plot on slide 4 shows plenty of pairs of people in the z1 20%-50% range. Nemesh also notes that some particular samples appear to be contaminated with other people’s DNA and thus exhibit elevated heterozygosity and cryptic relatedness to a large number of individuals. I’ll return to this point later.
Although it’s not perfect, for now I’m satisfied that my analysis validated the annotated (and unannotated but noticed-by-others) relationships in 1000 Genomes, and therefore is likely to discover any relationships among my own samples.
4. Sex validation
While I was comparing my data to the SNP array data in Step 2, I noticed that the pedigree files disagreed about the sex of one person. I decided it would be worthwhile to validate the sex of all individuals in my study based on the genetic data itself. I wrote a simple query to get the X het/hom ratio (an indicator of femaleness) and number of Y variants called (an indicator of maleness) for all of my samples. You might expect X heterozygosity to be 0 for men, and Y variants to be 0 for women, but in practice the pseudoautosomal region muddles this a bit. This region is homologous on X and Y, and so reads for women will often align to Y and reads from men’s Y chromosomes will often align to X. Still, these measures create pretty clear clusters of men and women.
select s.sid,
s.sex, -- nominal sex as annotated in my metadata
sum(case when chrom = 'X' and variant_allele_count=1 then 1.0 else 0.0 end)/sum(case when chrom = 'X' and variant_allele_count =2 then 1.0 else 0.00001 end) x_het_hom_ratio,
sum(case when chrom='Y' and sv.dp <> 'NA' then 1.0 else 0.0 end) y_vars
from sample_variants sv, variants v, samples s
where sv.vid = v.vid and s.sid = sv.sid and chrom in ('X','Y')
and sv.dp <> 'NA'
group by s.sid, s.sex
order by 3,4
;
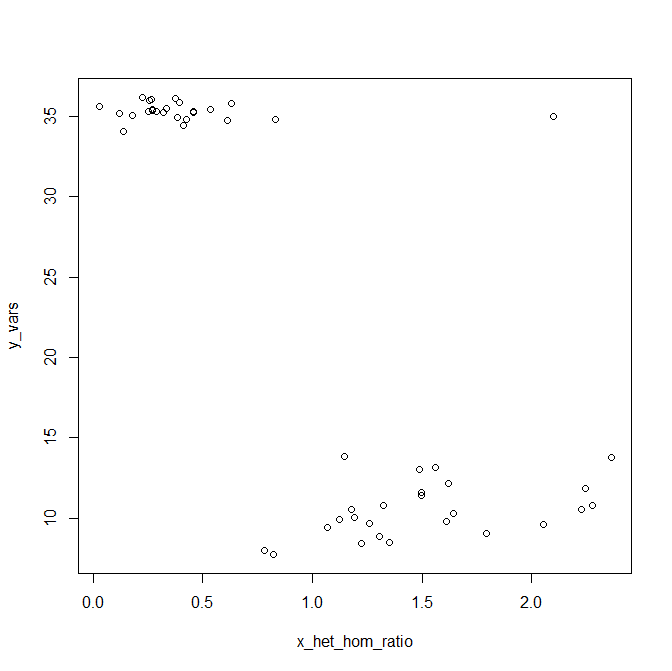
When I plotted these data, I found two really clear clusters: men with ~35 Y variants and an X het/hom ratio of about 0.5, and women with ~9 Y variants called and X het/hom ratio in the range of 1.3 to 2.2. But there was one outlier, a sample with 35 Y variants and an X het/hom ratio of 1.7. Here is some data I generated in R that looks about like my real data did (for various reasons it would be uncouth for me to post any real data on the blog):
y_vars = c(rnorm(m=35.32,sd=.5,25),rnorm(m=10,sd=2,25))
x_het_hom_ratio = c(rnorm(m=.34,sd=.2,25),rnorm(m=1.64,sd=.4,25))
y_vars[1] = 35
x_het_hom_ratio[1] = 2.1
plot(x_het_hom_ratio,y_vars)
At first I was certain this must be a Kleinfelter’s (XXY) individual. But after spending some quality time with the IBD figures from step 3, I started to get suspicious– this same individual also exhibited cryptic relatedness to a number of different 1000 Genomes people, including people from different subpopulations. When I read slide 8 of Jim Nemesh’s powerpoint, I started to understand why:
- Five samples have low-level reported “IBD” to dozens of other samples
- This can arise when a DNA sample or cell line has some contamination from another genome
- Three of these individuals also have excess heterozygosity
So then I calculated the exome-wide het/hom ratio for all of my samples:
select sid, sum(case when variant_allele_count = 1 then 1.0 else 0.0 end)/sum(case when variant_allele_count = 2 then 1.0 else 0.0 end) as het_hom_ratio
from sample_variants sv
group by sid
order by sid
;
And that clinched it. Virtually everyone had heterozygosity of about 1.7, but the one sample I had thought was XXY had a ratio of 3.4. Could it be an outlandish coincidence, that this one person had an XXY karyotype and was the only mixed-ancestry person in the sample? Another look at the IBD figures pretty much ruled this out: all the 1000 Genomes people to whom this individual was cryptically related were of the same race, which wouldn’t support the idea that this elevated heterozygosity was the result of mixed ancestry. So the parsimonious explanation was clear: this sample contains DNA from at least two different people, including at least one female and one male. Scanning the list of IBD pairs, it’s not clear which (if any) of my other samples was the source of contamination– several of them share high IBD with this sample. It looks like I have to simply exclude this sample from my study.
Having gone through this process for this one contaminated sample helps me understand why heterozygosity is among the quality metrics usually computed for variant data, and why cryptic relatedness can be a sign of more than just substructure you need to control for.
In fact, I went back to my original IBD table and tried excluding my one contaminated sample as well as HG00098 (which Nemesh notes as being possibly contaminated) and HG00124 (which I had noticed exhibited cryptic relatedness to tens of the 1000G Europeans in my dataset). Once these three samples were filtered out, the IBD data looked vastly more rational– after the familial relationships mentioned above and in my own data, the IBD figures dropped off pretty quickly to IBD-1 of 40% or less and a pi_hat of no more than about .25.
update 2012-11-12: when you calculate exome-wide heterozygosity, you’ll probably want to color-code the population each person belongs to, since homozygosity varies by population. Here’s some R code to make a colored bar chart:
library(RPostgreSQL) # first load package RPostgresql
drv <- dbDriver("PostgreSQL")
readcon <- dbConnect(drv, dbname="mydb", user="postgres", password="password")
sql <- "
select sv.sid, c.cluster, sum(case when sv.variant_allele_count = 1 then 1 else 0 end)::numeric/sum(case when sv.variant_allele_count = 2 then 1 else 0 end)::numeric as het_hom_ratio
from clusters c, sample_variants sv
where c.iid::integer = sv.sid
group by sv.sid, c.cluster
order by sv.sid
;
"
rs <- dbSendQuery(readcon,sql)
tbl <- fetch(rs,n=-1)
library(hash)
pops = c("African","Asian","European")
colors = c("forestgreen","blue","orange")
k = hash(keys=pops,values=colors)
kvec = tbl$cluster
for (i in 1:length(tbl$cluster)) { kvec[i] <- k[[tbl$cluster[i]]] }
barplot(tbl$het_hom_ratio,col=kvec,ylim=c(0,4),ylab="Exome-wide het/hom ratio",xlab="Samples",border=NA)
legend("topleft",pops,col=colors,cex=1.0,bty="n",pch=15)
dbDisconnect(readcon)
dbUnloadDriver(drv)
And the results (this is fake data that looks approximately like my real data):
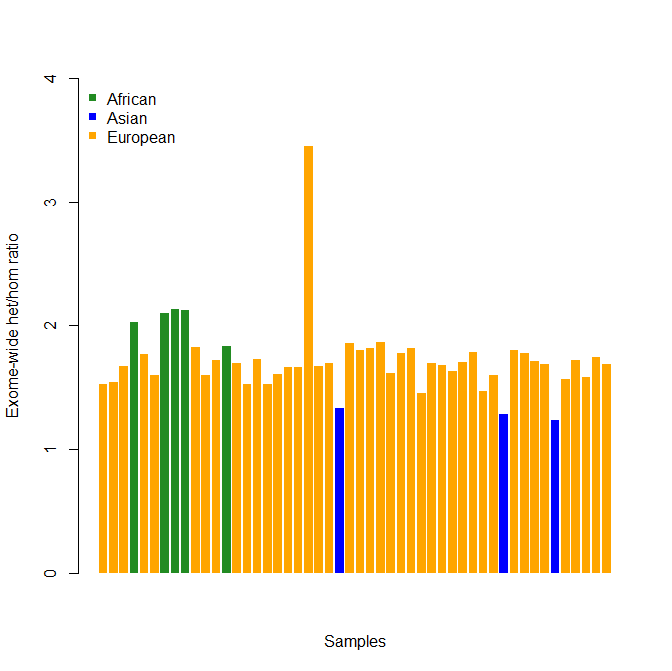
This makes it extremely clear which sample is elevated above baseline. That’s not a guarantee that sample is contaminated—you should still look at your MDS plots and IBD tables before you rule out the possibility the person is of mixed ancestry and is genuinely that heterozygous.