Chlorpromazine: a new mechanism of action
Way back in 2012, I wrote this post chronicling the life and (I thought) death of chlorpromazine as a candidate antiprion drug. The Prusiner lab had discovered that this FDA-approved antipsychotic drug inhibited the formation of proteinase K-resistant PrPSc in cell culture, with an EC50 of about 3 μM [Korth 2001], and this was validated by subsequent work in the Caughey lab [Kocisko 2003]. The drug went on to be tested, ad hoc, in a few patients on a compassionate use basis, with no clear evidence for efficacy (see post).
The story turns out to be more interesting than I thought. Baral 2014 reports that chlorpromazine binds to a hydrophobic pocket in PrPC nestled in between β-sheets 1 and 2 on one side and α-helix 2 on the other side. Its unchlorinated sister compound, promazine, binds in a nearly identical fashion. This was discovered through X-ray crystallography of MoPrP bound to the POM1 Fab and to promazine or chlorpromazine. Although the experiments used MoPrP, the paper uses HuPrP numbering and I’ll refer to HuPrP numbering and amino acids in this post.
The binding of chlorpromazine did not dramatically alter PrPC’s conformation, but its binding did engender multiple subtle conformational changes:
- It re-orients amino acids 117-124 (AAGAVVGG in HuPrP) into a third β-sheet (I’ll call it β0), whose backbone hydrogen bonds with that of β1.
- It stabilizes the β2-α2 loop. The exact conformation of this loop differs in different reported structures of PrPC, suggesting that this region has some conformational flexibility in unbound PrPC. When PrPC is bound to promazine, residues D178, Y128 and Y169 are stably hydrogen-bonded to each other.
- It reduces the conformational flexibility of α2 and α3 through allosteric effects secondary to the above changes.
- It serves to attach the β1-α1-β2 portion of PrPC to the face of α2. This may prevent the “unhinging” of these two subdomains, an event which has been proposed to be essential for prion conversion.
The main reason that all of this is interesting is that it suggests that perhaps chlorpromazine inhibits prion formation by binding PrPC. I had always gotten the impression that it was nearly impossible to find small molcules that bind PrPC. For instance, Poncet-Montange 2011 reported that 0 of 16 validated hits from an antiprion compound screen bound PrPC - instead, they all acted through indirect mechanisms. MRC Prion Unit specifically designed an assay to find compounds that bind PrPC, and yet the two hits for which John Collinge presented in vivo efficacy data at CJD2014 both have a “unique mechanism of action” and do not bind PrP.
Binding to PrPC is desirable because one might speculate that a compound with such a mechanism would have antiprion effects against all different strains of prions. Consider, for contrast, quinacrine. Like chlorpromazine, quinacrine reduces PK-resistant PrPSc formation in cultured cells. It is more potent, with an EC50 of about 300 nM. When quinacrine was conjugated to beads in an affinity chromatography matrix, it pulled down PrPSc much moreso than PrPC [Phuan 2007], implying that its inhibitory mechanism probably involves binding PrPSc. PrPSc comes in different conformations and can evolve and change — accordingly, quinacrine has only transient inhibitory effects and quickly gives rise to drug resistant prion strains [Ghaemmaghami & Ahn 2009]. Similar drug resistance and strain specificity problems have been observed with 2-aminothiazoles [Berry 2013] and so I would speculate that they bind PrPSc as well, though their mechanism is still unknown [Ghaemmaghami 2014].
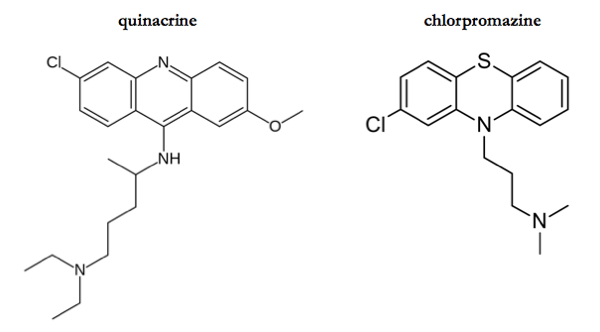
What is hard to believe is that chlorpromazine and quinacrine inhibit prions via completely different mechanisms. The two compounds are not so dissimilar (see above) - indeed, [Korth 2001] calls quinacrine “a structural antecedent” of phenothiazines such as chlorpromazine. While they of course each have their own targets in other realms of biology (quinacrine is an antimalarial of apparently unknown mechanism; chlorpromazine is a dopamine antagonist), to suppose that the two compounds inhibit prion formation by two entirely distinct mechanisms seems unparsimonious. Interestingly, the study of quinacrine’s mechanism that I have just mentioned [Phuan 2007] also tested the affinity of bead-conjugated chlorpromazine supplement. Chlorpromazine, too, pulled down PrPSc, though with only one-quarter the affinity of quinacrine (Table S3). Was chlorpromazine, then, binding PrPC in the same pocket as reported by [Baral 2014] and thereby indirectly pulling down PrPSc that was bound to that PrPC?? Or was chlorpromazine binding to PrPSc itself, as we suppose quinacrine does? Arguably, as you get into the details, the distinction between PrPC and PrPSc ceases to be black-and-white, as there may be a number of partially folded intermediates involved in the conversion pathway [Stohr 2008].
There are yet other pieces of evidence for different mechanisms too. I found one study [Vogtherr 2003] claiming yet a third possible mechanism / binding site for these compounds. Vogtherr provided NMR evidence for quinacrine binding to the extreme C-terminal end of PrPC (residues 225-227). However, this was observed at millimolar affinity, whereas quinacrine inhibits PK-resistant PrPSc accumulation with a 300 nM EC50. At those millimolar concentrations, that study also found that chlorpromazine bound to PrPC at the same site. One new cell biology study, just out yesterday [Yamasaki 2014] reports that chlorpromazine changes the subcellular distribution and metabolism of PrPSc but not of PrPC.
The question of chlorpromazine’s inhibitory mechanism seems important. Baral ends on a therapeutic-oriented note:
Our high-resolution structural data will be very useful toward enhancing the therapeutic potential of tricyclic compounds through a structure-guided approach.
And indeed, if chlorpromazine really does bind PrPC, then it would seem prudent to prioritize defining its structure-activity relationship and improving its antiprion properties while abolishing its dopamine antagonist properties.
All this made me wonder, do we actually know whether chlorpromazine has efficacy in vivo? As mentioned above, there were a few ad hoc compassionate use applications in patients, but their failure could be chalked up to any number of different explanations: was it late administration, strain specificity, insufficient concentrations in the brain, inability to inhibit prions in vivo, or simply the total lack of any statistical power in these ad hoc studies? An in vivo efficacy study would go much further toward determining whether chlorpromazine “works” than these trials in patients did.
And in fact, chlorpromazine has been tested in scrapie-infected mice once, in a volume so ancient and obscure that when I wrote my earlier post I wasn’t able to actually track the article down, only the abstract [Roikhel 1984]. After getting my new student ID last week, I ventured into the bowels of Countway Library and retrieved the full text. It seems that the authors treated each mouse once weekly with 0.2 mL of an 0.25% (presumably wt/vol, i.e. g/mL) solution of chlorpromazine (then called aminasamine). That works out to 0.5 mg of drug, which for (say) a 30g mouse is 17 mg/kg, a decent test dose, though the drug would have to have some amazing pharmacokinetic properties to stay in the brain for a whole week. The paper doesn’t say how often the mice were checked for scrapie signs, but my guess would be only once per month, based on the fact that the onsets for various groups are reported in units of 30 days post infection (150, 180 and 210 dpi) even though there were ≥20 mice in each group.
In short, this old study was not exactly done to the standards of today’s prion science. I would consider it an entirely open question whether chlorpromazine “works” in vivo.
Given its potentially interesting mechanism, its known efficacy in cell culture and its entirely unknown performance in vivo against prion infection, I’m prepared to revise my verdict on chlorpromazine from “another prion therapeutic of yesteryear” to a potentially interesting lead compound that merits some further study. I am incredibly inexpert at both structural biology and medicinal chemistry, though, so I would encourage readers who know more about these domains to leave some comments below to help me understand the implications of Baral’s work.
